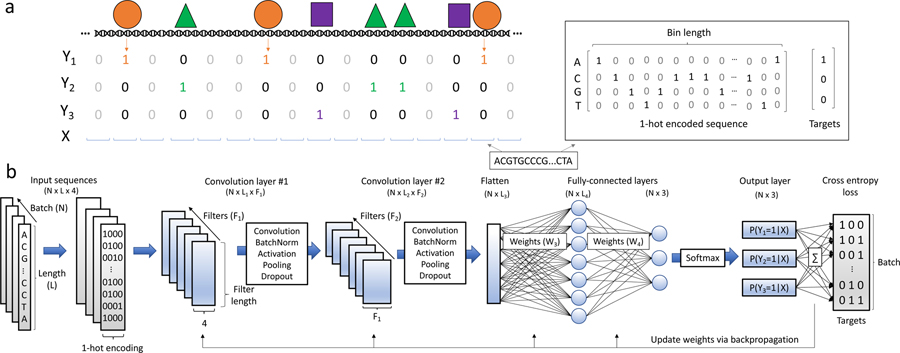
Figure 1:
Overview of TF binding site prediction task. a) Transcription factors bind to regions of the genome based on sequence specificities and modulate various biological functions. ChIP-seq experiments enrich for short DNA sequences that are interacting with the TF under investigation. The resultant DNA sequences (so-called reads) are aligned to a reference genome and a peak calling tool is employed to find read distributions that are statistically significant compared to background levels. Upon binning the full genome into bins of length L, it is possible to then associate each bin with a binary label denoting the presence (Yi = 1) or absence (Yi = 0) of TF i based on sufficient overlap between the peaks and the bin. The DNA within each bin is represented by a 1-hot encoded matrix and the associated label vectors are used to train a model as a single-class or multi-class supervised learning task. b) Convolutional neural networks are powerful methods to learn sequence-function relationships directly from DNA sequence. A CNN is comprised of a number of first layer filters (F1) which learn features directly from the N input sequences by computing the cross-correlation between each set of filter weights and the 1-hot encoded sequence. The resultant scans, so-called feature maps, intuitively represent the match between each pattern being learned in a given filter and the input sequence. The feature map then undergoes a series of functional (e.g. batch normalization, non-linear activation) and spatial transformations (e.g. pooling) resulting in a truncated length (L1). This tensor is then fed into deeper convolutional layers which discriminate higher-order relationships between the learned features. Two convolutional blocks are depicted however this feed-forward process may be repeated any number of times, after which a flattening operation is utilized to reshape the tensor into a N × L3 matrix. Fully-connected layers perform additional matrix multiplications and ultimately output a probability of class membership for each target. Loss is calculated between the predicted values and the targets, and the weights are updated with a learning rule that uses backpropagation to calculate gradients throughout network.