Figure 4.
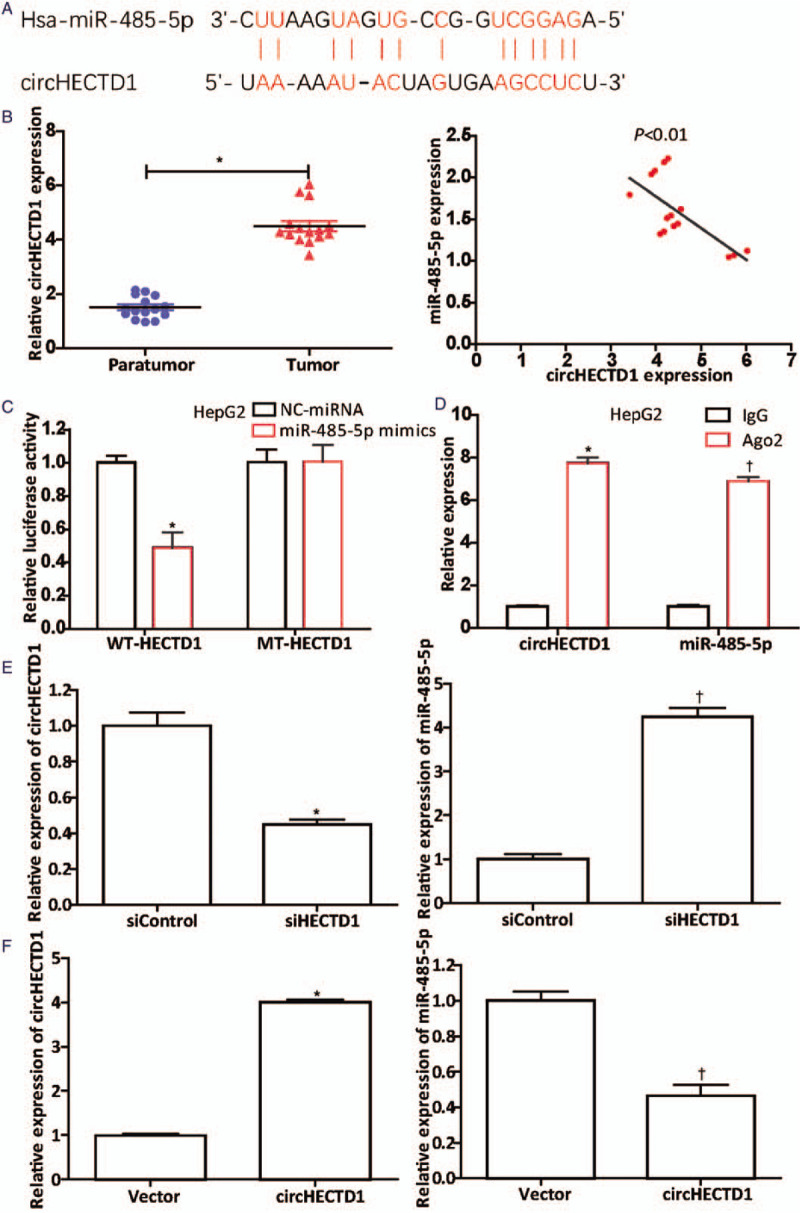
CircHECTD1 acts as a miR-485-5p sponge. (A) Complementary sequences between circHECTD1 and miR-485-5p discovered from the Starbase 2.0 database analysis. (B) qRT-PCR showed that circHECTD1 in HCC tissues (n = 15) was dramatically up-regulated. ∗P < 0.001 compared with the level in paratumor tissues. The correlation between their expression levels was negative (P < 0.01). (C) Luciferase activity was detected after cotransfection of WT-HECTD1- or MT-HECTD1-containing luciferase reporter gene with miR-485-5p mimics or NC-miRNA into HepG2 cells, according to the Lipofectamine 3000 manufacturer's instructions. ∗P < 0.01 compared with NC-miRNA group. (D) The circHECTD1 and miR-485-5p content in Ago2 pellets was verified using the RIP assay. ∗P < 0.001 (circHECTD1) compared with that in IgG pellets; †P < 0.001 (miR-485-5p) compared with that in IgG pellets. Further, qRT-PCR was conducted to investigate the effects of (E) down-regulation (circHECTD1: ∗P < 0.01 compared with the siControl group; miR-485-5p: †P < 0.001 compared with the siControl group) or (F) up-regulation of circHECTD1 (circHECTD1: ∗P < 0.001 compared with the vector; miR-485-5p: †P < 0.01 compared with the vector) on the miR-485-5p level. The data in (C–F) are presented as mean ± standard deviation (n = 3). HCC: Hepatocellular carcinoma; MiR: MicroRNA; MT-HECTD1: Mutant-type HECTD1; qRT-PCR: Quantitative real-time polymerase chain reaction; RIP: RNA immunoprecipitation; WT-HECTD1: Wild-type HECTD1.
