Table 3. Data-collection and refinement statistics for HDAC6 CD1 mutant–inhibitor complexes.
Values in parentheses are for the highest resolution shell.
| HDAC6 CD1 mutant | H82F/F202Y | K330L | |
|---|---|---|---|
| Inhibitor | Trichostatin A | SAHA-BPyne | 4-Iodo-SAHA |
| Data collection | |||
| Beamline | 24-ID-C, APS | 17-ID-1, NSLS-II | 24-ID-E, APS |
| Wavelength (Å) | 0.98 | 0.92 | 0.98 |
| Temperature (K) | 100 | 100 | 100 |
| Detector | PILATUS 6M-F | EIGER 9M | EIGER 16M |
| Wilson B factor (Å2) | 32 | 46 | 15 |
| Crystal-to-detector distance (mm) | 300 | 230 | 150 |
| Rotation range per image (°) | 0.20 | 0.20 | 0.20 |
| Total rotation range (°) | 180 | 180 | 180 |
| Exposure time per image (s) | 0.20 | 0.02 | 0.20 |
| Space group | P21 | C22121 | P21 |
| a, b, c (Å) | 53.1, 124.0, 55.0 | 66.0, 95.2, 119.7 | 53.1, 123.8, 55.1 |
| α, β, γ (°) | 90.0, 114.4, 90.0 | 90.0, 90.0, 90.0 | 90.0, 113.5, 90.0 |
| R merge † | 0.054 (0.418) | 0.190 (0.817) | 0.091 (0.271) |
| R p.i.m. ‡ | 0.052 (0.395) | 0.119 (0.507) | 0.071 (0.231) |
| CC1/2 § | 0.996 (0.792) | 0.971 (0.618) | 0.981 (0.829) |
| Multiplicity | 3.4 (3.2) | 6.4 (6.7) | 2.4 (2.3) |
| Completeness (%) | 98.7 (98.4) | 100.0 (100.0) | 77.9 (80.6) |
| 〈I/σ(I)〉 | 8.9 (1.9) | 4.8 (2.0) | 5.2 (2.0) |
| Refinement | |||
| Resolution (Å) | 48.34–2.30 (2.38–2.30) | 54.25–2.40 (2.49–2.40) | 39.14–1.90 (1.97–1.90) |
| No. of reflections | 28350 (2842) | 15104 (1483) | 39267 (3884) |
| R work/R free ¶ | 0.170/0.232 (0.210/0.256) | 0.204/0.243 (0.263/0.274) | 0.177/0.223 (0.206/0.270) |
| No. of atoms†† | |||
| Protein | 5288 | 2688 | 5338 |
| Ligand | 50 | 73 | 46 |
| Solvent | 116 | 47 | 268 |
| Average B factors (Å2) | |||
| Protein | 33 | 47 | 14 |
| Ligand | 29 | 44 | 17 |
| Solvent | 31 | 43 | 18 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.007 | 0.002 | 0.007 |
| Bond angles (°) | 0.9 | 0.6 | 0.8 |
| Ramachandran plot‡‡ | |||
| Favored | 95.53 | 96.63 | 96.30 |
| Allowed | 4.47 | 3.37 | 3.70 |
| Outliers | 0.00 | 0.00 | 0.00 |
| PDB code | 6wyo | 6wyp | 6wyq |
R
merge = 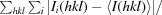
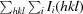 , where 〈I(hkl)〉 is the average intensity calculated for reflection hkl from replicate measurements.
, where 〈I(hkl)〉 is the average intensity calculated for reflection hkl from replicate measurements.
R
p.i.m. = 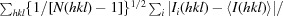
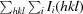 , where 〈I(hkl)〉 is the average intensity calculated for reflection hkl from replicate measurements and N(hkl) is the number of reflections.
, where 〈I(hkl)〉 is the average intensity calculated for reflection hkl from replicate measurements and N(hkl) is the number of reflections.
Pearson correlation coefficient between random half data sets.
R
work = 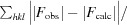
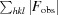 for reflections contained in the working set. |F
obs| and |F
calc| are the observed and calculated structure-factor amplitudes, respectively. R
free is calculated using the same expression for reflections contained in the test set that was held aside during refinement.
for reflections contained in the working set. |F
obs| and |F
calc| are the observed and calculated structure-factor amplitudes, respectively. R
free is calculated using the same expression for reflections contained in the test set that was held aside during refinement.
Per asymmetric unit.
Calculated with MolProbity.
