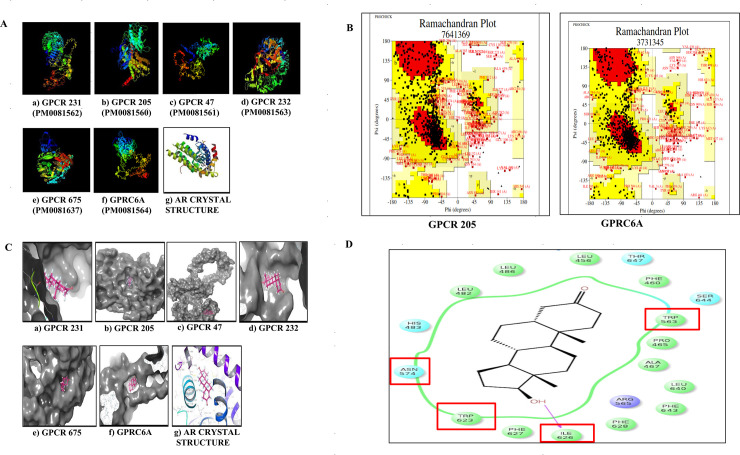
Fig 1. Sequence comparison of mammalian GPCRs and the ligand binding domain of AR.
A) The 3D structures of candidate GPCRs as predicted using I-TASSER. The 3D models of the target protein were deposited to the Protein Model Database (PMDB) and each model was assigned a unique PMDB model identifier ID. B) Example of Ramachandran plots for selection of best model of the GPCRs (i.e. for GPRC6A and GPCR 205 (Ramachandran plot for other GPCRs not shown). The models for the GPCRs were selected based on the maximum residues obtained in the most favored region (MFR) using PROCHECK software. C) In silico molecular docking of selected GPCRs with testosterone (ligand) using Schrodinger Glide Maestro. Using multiple sequence alignment (clustal W) the conserved regions in selected GPCRs were identified. Testosterone was docked at the conserved region of the target GPCR to have a specific region docking prediction. D) Residue interaction diagram using Schrodinger Maestro software showing residues surrounding testosterone. The critical residues capable of making covalent interactions with testosterone are I626, W623, W563, N574. The residues I626 and W623 were mutated into Proline and Arginine using site directed mutagenesis for mutation experiments.