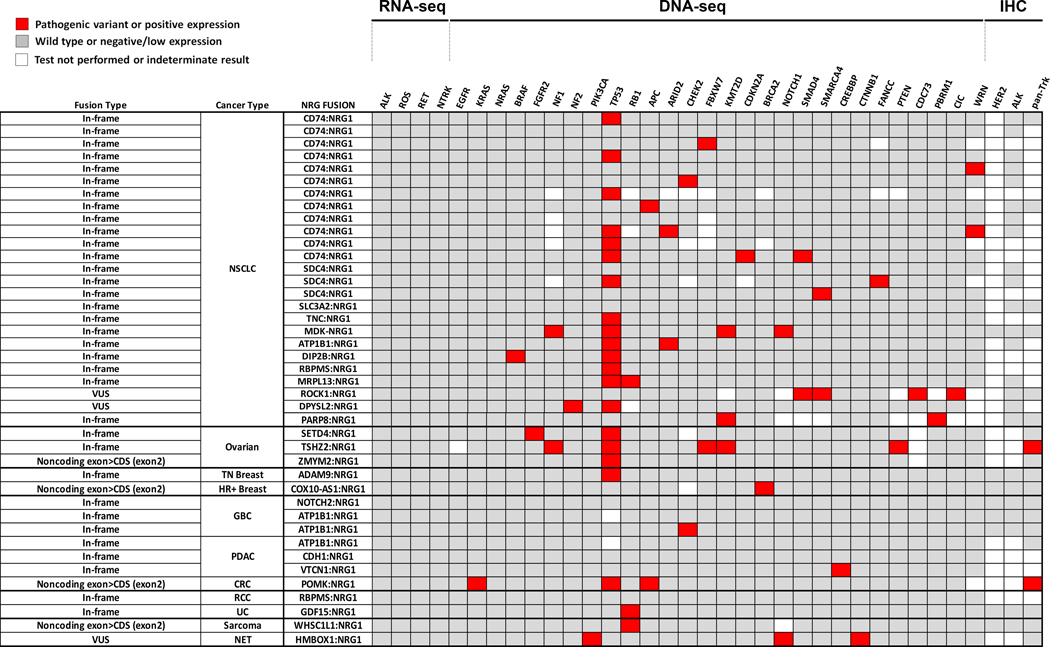
Figure 2.
Genomic features observed in NRG1-fusion positive solid tumors. Oncoprint plot illustrating co-occurrence of driver events, genes with any pathogenic variant detected in the cohort and other clinically relevant protein markers. Each NRG1-fusion positive sample corresponds to one row in the table: frame prediction of the fusion, cancer type and fusion partner are provided. Fill of boxes correlate with gene/protein status: (1) red, pathogenic variant detected or positive expression, (2) grey, wild type or low/negative expression and (3) white, test was not performed, or indeterminate. Pathogenic variants in oncogenes were rare, but at least one mutation in tumor suppressor genes including TP53 occurred in all but nine samples. Abbreviations: NSCLC, non-small cell lung cancer, TN (triple-negative) breast, HR+ (hormone receptor positive) breast, GBC, gallbladder cancer (cholangiocarcinoma); PDAC, pancreatic ductal adenocarcinoma; CRC, colorectal cancer; RCC, renal cell carcinoma; UC (urothelial bladder cancer); NET (neuroendocrine tumor of the nasopharynx); VUS, fusion variant of unknown significance; CDS, coding sequence.