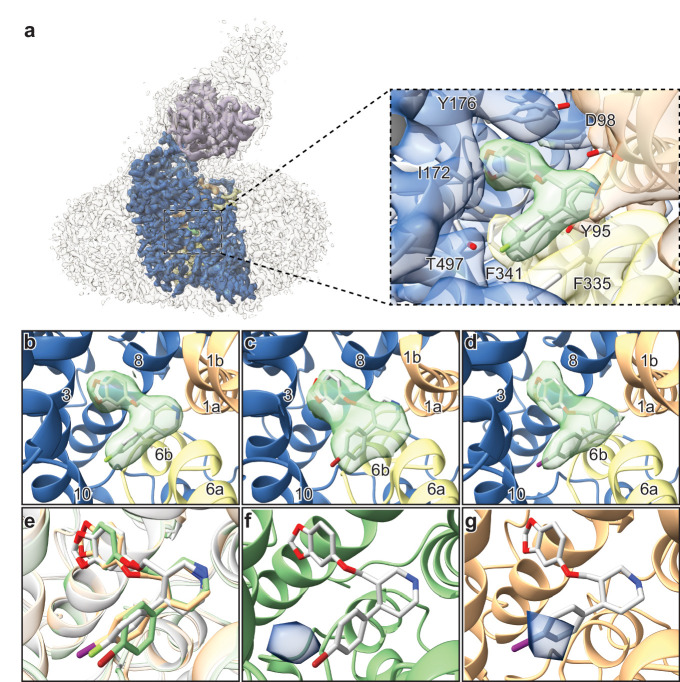
Figure 4. Structures of SERT-paroxetine complexes.
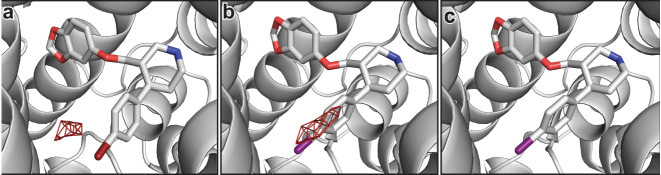
(a) Cryo-EM reconstruction of SERT bound to paroxetine where the shape of the SERT-8B6 Fab complex and detergent micelle is shown in transparent light grey. The density of SERT is shown in dark blue with TM1 and TM6 colored in orange and yellow, respectively, and the density for paroxetine in green. The variable domain of the 8B6 Fab is colored in purple. Inset shows the density features at the central site of paroxetine. (b) Density feature at the central site of paroxetine. (c) Density feature at the central site of Br-paroxetine. (d) Density feature at the central site of I-paroxetine. (e) Comparison of the binding poses of paroxetine (grey), Br-paroxetine (green), and I-paroxetine (orange). (f) Anomalous difference electron density (blue) derived from Br-paroxetine, contoured at 5.2σ. g, Anomalous difference electron density (blue) derived from I-paroxetine, contoured at 4.3σ.
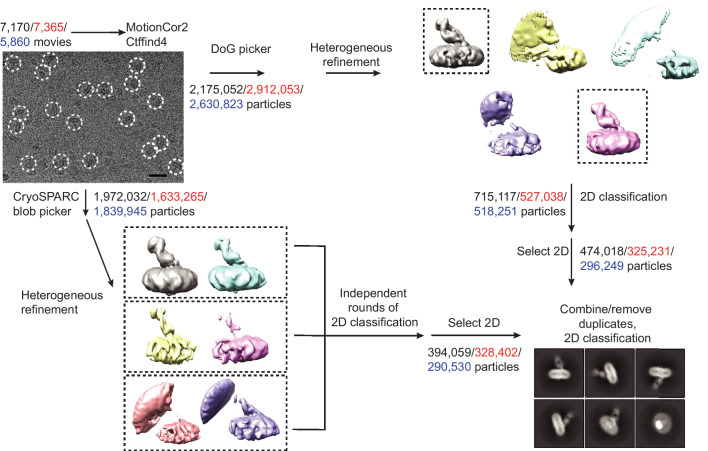
Figure 4—figure supplement 1. Work-flow of cryo-EM data processing of ΔN72/ΔC13 SERT/8B6 Fab/paroxetine complexes.
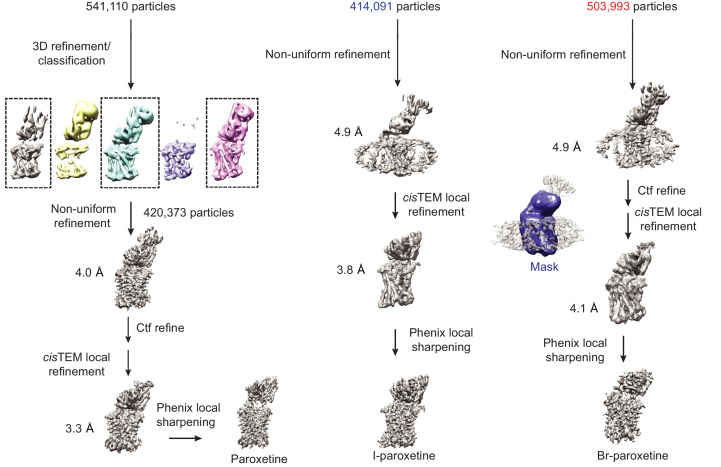
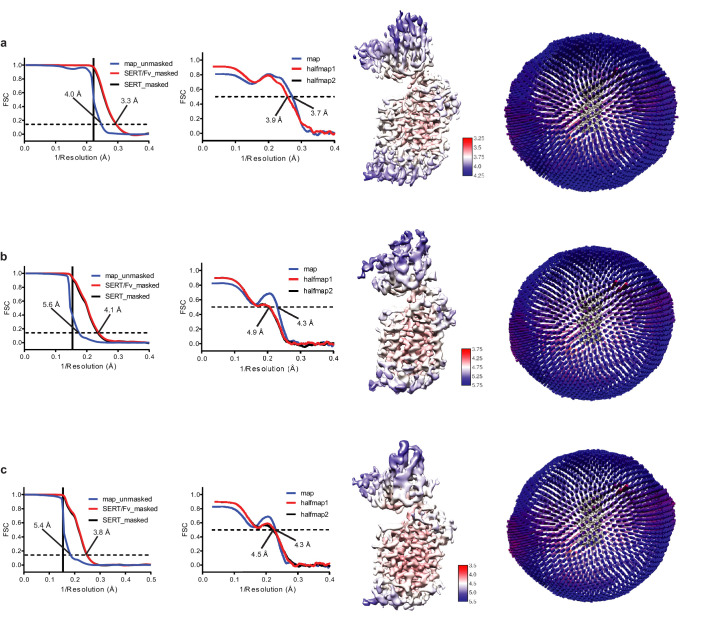
Figure 4—figure supplement 2. 3D refinement of ΔN72/ΔC13 SERT/8B6 Fab/paroxetine complexes.
Figure 4—figure supplement 3. Cryo-EM reconstruction of ΔN72/ΔC13 SERT/8B6 Fab/paroxetine complexes.
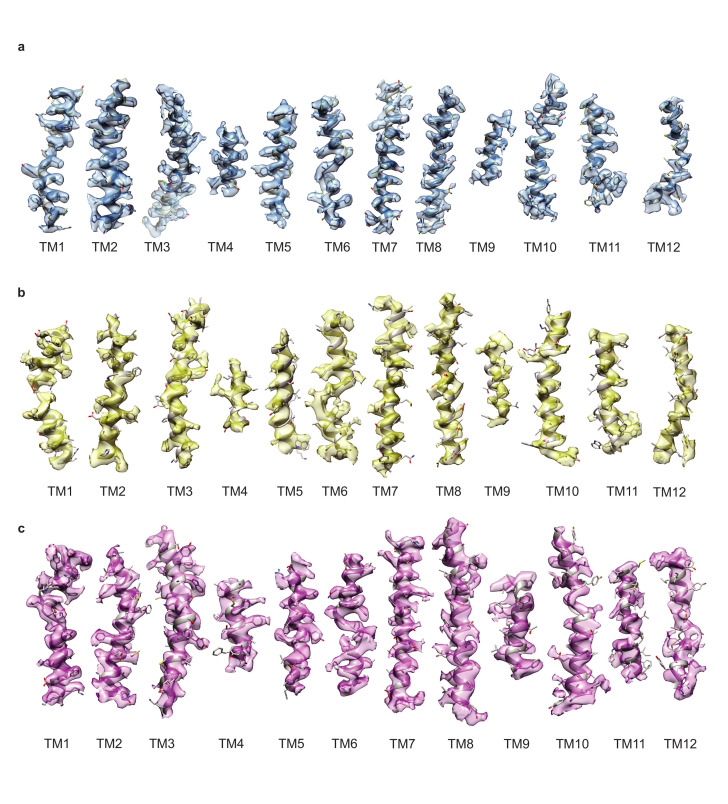
Figure 4—figure supplement 4. Cryo-EM density segments of the transmembrane helices.
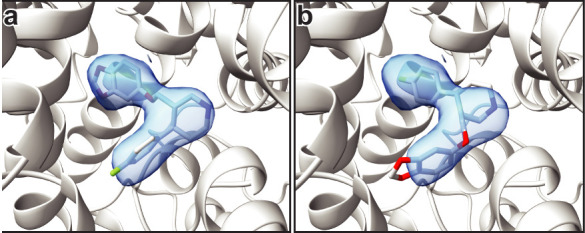
Figure 4—figure supplement 5. Comparison of the fit of paroxetine in the ABC and ACB poses.