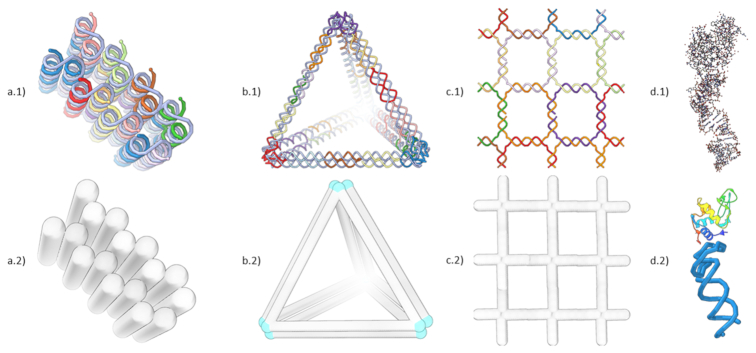
Figure 1.
Screenshots of different DNA nanostructure design concepts and their visualization in Adenita. (a) A multi-layer DNA brick designed with Cadnano, (b) a tetrahedron designed using Daedalus and (c) a squared lattice DNA Tile that was manually designed. In Cadnano structures, all double helices must be parallel, while DNA wireframe approaches place double strands at the edges of a mesh. DNA tiles require the repetition of one or several small DNA motifs to create a 2D or 3D shape. In this example, we used a four-arm Holliday junction. (d.1) shows the default all-atom model of the PDB structure 4M4O, formed by a protein and an aptamer, in (d.2) a combination of Adenita's visual model and Samson's secondary structure visualization makes it possible to simplify the representation of such molecules.