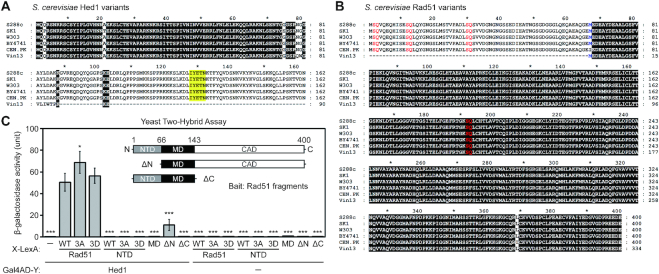
Figure 1.
Interactions of Rad51 and Hed1 proteins. Sequence comparison of Hed1 and Rad51 in different yeast strains reveals loss of the COOH-terminal region of Hed1 (A), and the NH2-terminal region of Rad51 (B) in the budding yeast strain Vin13. Protein sequences were retrieved from the Saccharomyces Genome Database (SGD). Multiple sequence alignments were performed with MAFFT version 7 (63), and the results were displayed and edited with GeneDoc (version 2.7.000; https://www.softpedia.com/get/Science-CAD/GeneDoc.shtml). Identical residues are shaded in black. (A) The reported Rad51-interacting region of Hed1 is shaded in yellow. (B) The initial 66 amino acid stretch (Met1 to Glu66) of S. cerevisiae Rad51 is defined as the NH2-terminal domain (NTD). Three SQ motifs (S2Q, S12Q and S30Q) in the NTD and one S192Q motif in the ATPase domain are highlighted in red. The methionine (Met67) following the NTD is shown in blue. (C) Yeast two-hybrid assay showing interactions between Rad51 fragments and Hed1. Quantitative yeast LacZ assays were performed as described previously (40,41). Hed1 is expressed from pGADT7 vectors (Clontech, USA), whereas in X-LexA the bait protein X is NH2-terminal to LexA (64) (Supplementary Table S3). Error bars indicate standard deviation between experiments (n ≥ 3, see Supplementary Table S3 for the exact sample sizes). Asterisks indicate significant differences when compared to values of interaction between WT Rad51–LexA and Gal4AD–Hed1, with P values calculated using a two-tailed t-test (*P value < 0.05 and ***P value < 0.001).