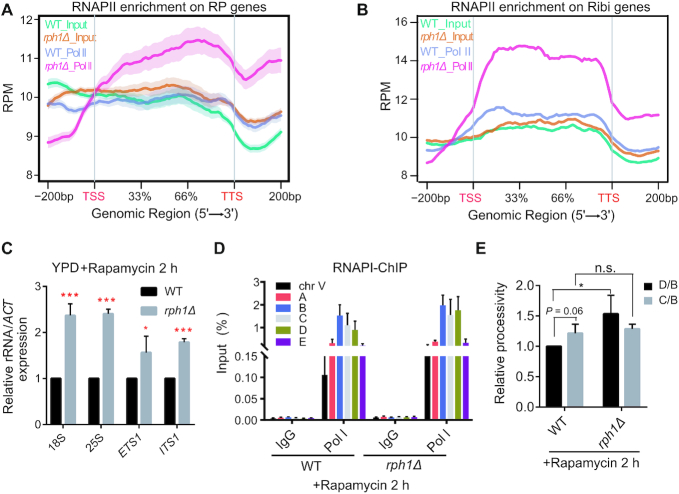
Figure 5.
Deletion of Rph1 increased RNAPII occupancy and facilitated RNAPI transcription with rapamycin treatment. (A and B) Distribution of RNAPII ChIP-seq signals on gene bodies of RPGs (A) and Ribi genes (B) in WT and rph1Δ cells with rapamycin treatment. (C) Expression of the indicated rRNA regions in WT and rph1Δ cells. (D) ChIP-qPCR analysis were performed to examine RNAPI (Rpa190-Flag) enrichment over different rDNA loci in WT or rph1Δ cells. IgG ChIP served as a negative control. (E) Processivity assay of RNAPI transcription were performed in the rph1Δ cells relative to WT. The IP/Input value for the middle gene body (the C locus) or the 3′ end (the D locus) was divided by the value for the 5′ end (the B locus), and the ratio (‘processivity’) for rph1Δ was normalized to that for WT. qPCR data are represented as mean ± SD from three biological replicates. t-test, ∗P < 0.05; ∗∗P < 0.01; *** P < 0.001, n.s. ‘not significant’.