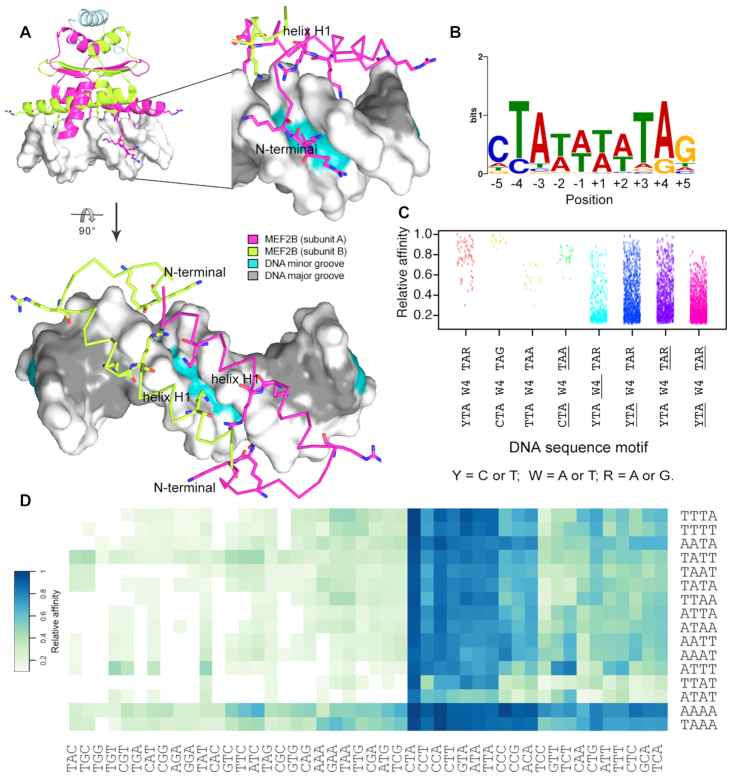
Figure 1.
Sequence variations for MEF2B binding sites. (A) Major MEF2B–DNA interactions based on co-crystal structure of MEF2B in complex with DNA (PDB ID: 1N6J) (27) are observed at the DNA minor groove and backbone. Amino acids that interact with DNA, including positively charged residues in the vicinity of DNA, are shown in stick representation. Different views are shown to depict binding recognition by helix H1 and N-terminal tail regions. (B) PWM obtained from SELEX-seq data using MEME Suite (45). Analyzed sequences were obtained after two rounds of selection for top 10-mers with relative affinity >0.7. (C) Strip chart showing relative binding affinities for 10-mers displaying full or partial matches to MEF2B consensus motif, represented by YTAW4TAR (Y = C or T; W = A or T; R = A or G). Variations in core motif are represented by underlined regions, with W4, YTA and TAR denoting regions that deviate from W4, YTA and TAR, respectively. (D) Heat map of relative affinities for triplet variations at 5′ peripheral half-site (YTAW4TAR) based on central core (W4) preferences.