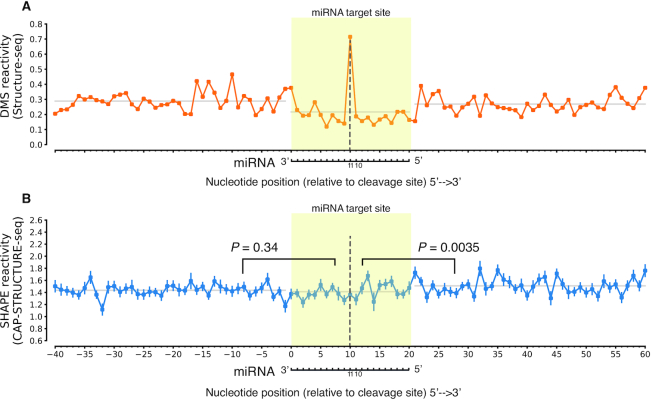
Figure 2.
CAP-STRUCTURE-seq can accurately probe intact mRNA structures in vivo. (A) Reactivity profiles across the miRNA target sites for the DMS reactivity from Structure-seq (8). The miRNA-mediated cleavage in the mRNA target site occurs at the tenth nucleotide of miRNA complementary sites, which leads to a skewed DMS reactivity profile at the eleventh nucleotide of miRNA complementary sites. Mean reactivity is shown. (B) SHAPE reactivity profiles from CAP-STRUCTURE-seq. The miRNA cleaved products do not show a peak at the cleavage sites, indicating the elimination of false positive signals. Mean reactivity together with SEM is shown. In A and B the yellow shading indicates the target binding sites. The dotted lines refer to the 11th nucleotide opposite to the miRNA. The horizontal axis is labeled from the 5′ end of the target gene to the 3′ end. P-values from one sided Mann–Whitney-U tests.