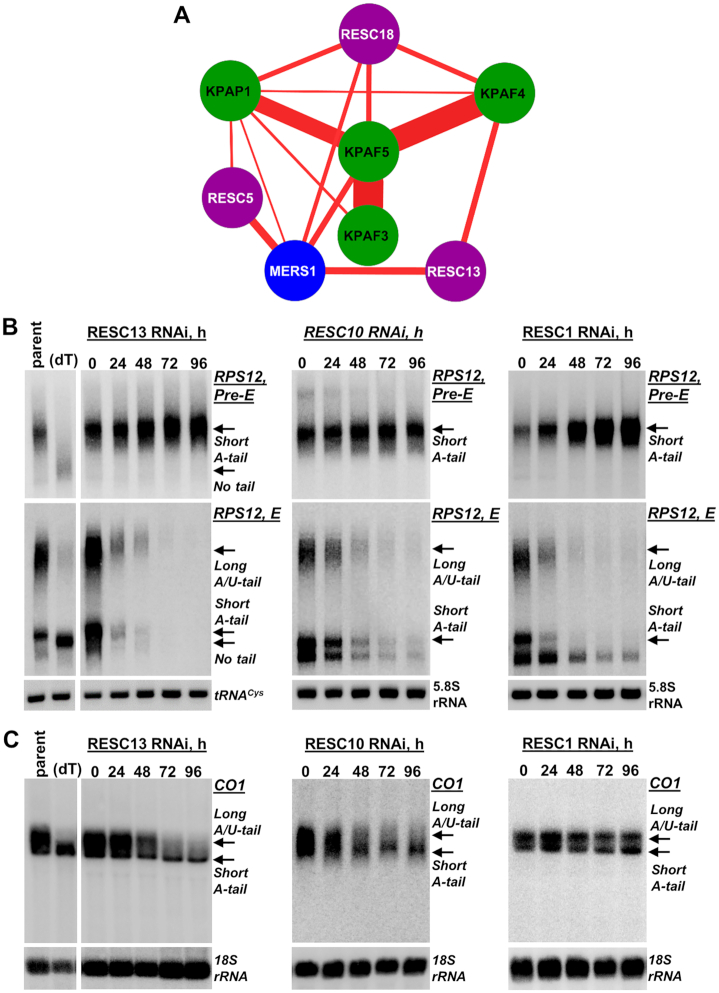
Figure 9.
RNA editing substrate binding complex (RESC) stabilizes never-edited mRNAs. (A) In vivo proximity network of KPAP1 poly(A) polymerase, KPAF3, 4 and 5, and RESC5, 13 (RGG2) and 18, and PPsome's pyrophosphohydrolase MERS1. The BioID experiments were performed in parallel under uniform induction, biotinylation, and purification conditions. The network was generated in Cytoscape software. The edge thickness correlates with normalized spectral abundance factor (NSAF) ranging from 1.13 × 10−4 (MERS1–KPAP1) to 1.5 × 10−3 (KPAF4–KPAF5) and reflects the predicted interaction strength (Supplementary Table S1). (B) Northern blotting of pre-edited (Pre-E) and fully-edited (E) RPS12 mRNA variants in RESC10, RESC13 (RGG2) and RESC1 RNAi knockdowns. Total RNA was separated on a 5% polyacrylamide/8M urea gel and sequentially hybridized with radiolabeled DNA probes. Parent: Lister 427 29–13 strain; (dT), RNA was hybridized with 20-mer oligo(dT) and treated with RNase H to remove A-tails and locate migration positions of a non-adenylated molecules. Short A-tailed and A/U-tailed mRNAs are indicated by arrows. Zero-time point: mock-induced RNAi cell line. Cytosolic 5.8S rRNA or tRNACys were used as loading control. (C) Northern blotting of never-edited CO1 mRNA in RESC10, RESC13 and RESC1 RNAi knockdowns. Total RNA was separated on a 1.7% agarose/formaldehyde gel and hybridized with oligonucleotide probe. Loading control: 18S rRNA.