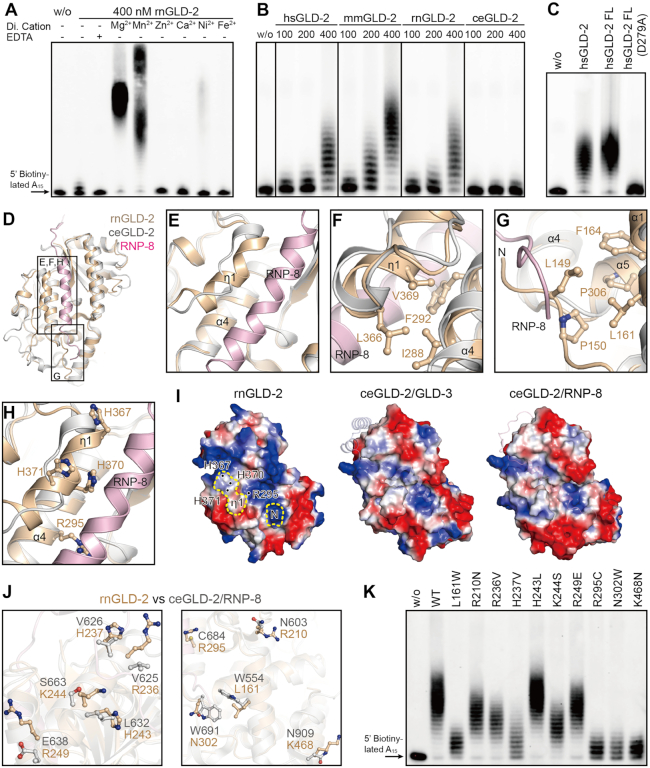
Figure 3.
rnGLD-2 is a potent PAP. (A) The PAP activity of rnGLD-2 in the presence of various divalent cations. A total of 400 nM rnGLD-2 was incubated with 500 nM 5′ biotinylated A15 RNA oligo and 500 μM ATP. For each sample, 2 mM indicated divalent ion or EDTA was supplied to the reaction. w/o, without protein. (B) Protein concentration-dependent PAP activity of hsGLD-2, mmGLD-2, rnGLD-2 and ceGLD-2. (C) Comparison between truncated and full-length hsGLD-2 in PAP activity. (D) Overall structural comparison between rnGLD-2 and ceGLD-2/RNP-8 complex from the backside of the catalytic domain, where the interaction site of ceGLD-2 and RNP-8 can be seen. Frames indicate the areas shown in panels (E–H). (E−H) Detailed structure differences between rnGLD-2 and ceGLD-2 explaining the discrepancy of their PAP activity. Color as in D. (E) The unique 310 helix (η1) of rnGLD-2 and its relative position to RNP-8. (F) Interaction between η1 and α4 buries the local hydrophobic cluster of rnGLD-2. (G) The N-terminal tip of rnGLD-2 shelters the hydrophobic residues on α1 and α5. (H) The histidine cluster on η1 and non-conserved Arg295 of rnGLD-2. (I) Electrostatic surface potential comparison at the backside of catalytic center for rnGLD-2 (left) and ceGLD-2 complexed with GLD-3 (middle) or RNP-8 (right). Locations of the N-terminal tip (N) and 310 helix (η1) of rnGLD-2 are outlined by yellow dashes, and the histidine cluster and Arg295 are indicated. GLD-3 and RNP-8 are shown in cartoon representation with 50% transparency. (J) Non-conserved surface residues between rnGLD-2 and ceGLD-2 that may affect substrate binding. (K) PAP activity of rnGLD-2 with mutations regarding the residue difference from ceGLD-2 as shown in (J).