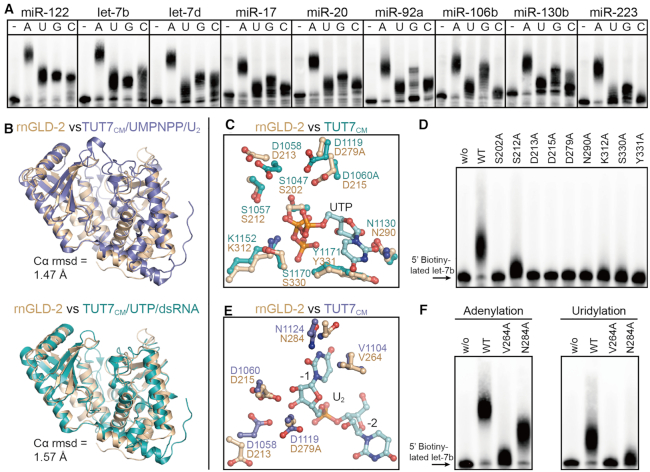
Figure 6.
Similarity between rnGLD-2 and TUT7CM. (A) NTase activity of rnGLD-2 on different miRNA substrates. A total of 400 nM rnGLD-2, 500 nM various 5′-biotinylated miRNA and 500 μM ATP/UTP/GTP/CTP were used. (B) Overall structure comparison between rnGLD-2 and TUT7CM in complex with UMPNPP/U2 (upper, PDB code: 5w0n) and with UTP/dsRNA (lower, PDB code: 5w0o). (C) Comparison between rnGLD-2 and TUT7CM at UTP binding site. Residues of TUT7CM (PDB code: 5w0o) involved in binding UTP is superimposed with corresponding residues of rnGLD-2. (D) Uridylation activity of rnGLD-2 with mutations regarding the residues potentially involved in UTP coordination. (E) Comparison between rnGLD-2 and TUT7CM at substrate binding site. Residues of TUT7CM (PDB code: 5w0n) involved in binding the U2 substrate is superimposed with corresponding residues of rnGLD-2. (F) Adenylation and uridylation activity of rnGLD-2 with mutations regarding the residues potentially involved in substrate binding.