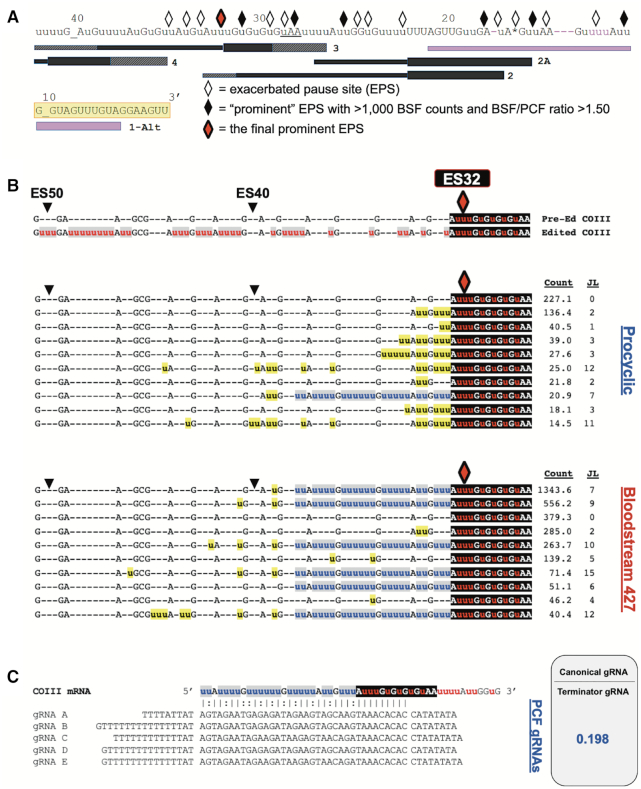
Figure 5.
Statistical and bioinformatic analyses of COIII mRNA editing intermediates at the final prominent EPS in PCF and BSF 427 cells. (A) Schematic illustrating the positions of exacerbated pause sites (EPSs) in BSF COIII mRNA in relation to the positions of gRNA families. gRNA anchor regions are illustrated as a box while the coding information region is illustrated with a line. The hatchmarked boxes at gRNA 5′ and 3′ ends represent variation in gRNA lengths within a gRNA family. The BSF 427 cells have an alternative editing consensus sequence that differs from the canonical sequence (shown in pink). The alternative guide RNA (1-Alt) that directs the editing of this alternative sequence is shown as a purple gRNA. Every 10 ESs are indicated above the sequence. Underscores at ES10 and ES40 are inserted to clarify the position of the ES. The yellow shaded regions indicate where the reverse COIII HTS primer anneals. The red diamond represents the final prominent EPS at ES32 in this cell line. (B) Sequence alignments of the 10 most abundant junction sequences at ES32 in COIII mRNA of PCF and BSF 427 cells. The canonical edited sequence is shown red and white letters with gray or black shading. Junction sequences are shown in black letters with yellow shading. Non-canonical consensus sequences are shown in blue letters with gray shading. To the right of each sequence is the average normalized read counts (Count) for the sequence and the junction length (JL). (C) Schematic of terminator guide RNAs found in PCF (gRNAs A–E) annealed to COIII mRNA with the non-productive terminator consensus junction sequence at ES32. The ratio of the counts for the canonical gRNAs that direct the editing past ES32 and the terminator gRNAs are shown as determined by previously published gRNA reads (61,62).