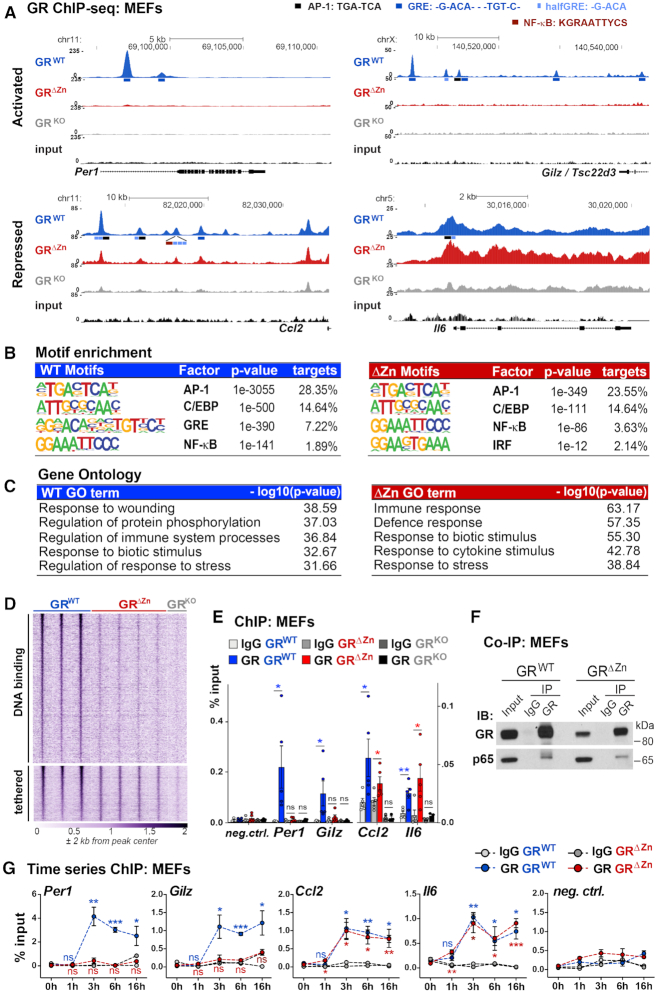
Figure 2.
Tethered binding sites are found near inflammatory genes in GRΔZn MEFs. (A) Representative GR ChIP-Seq tracks from wild type, homozygous mutant and knockout MEFs treated with LPS for 3 h and Dex overnight, with the positions of predicted motifs (not to scale!), (wild type n = 3, mutant n = 4 and knockout n = 2). (B) Enriched consensus motifs in GR ChIP-sequences. Known motifs were identified among wild type or mutant peaks. (C) Functional annotation of GR ChIP peaks assigned to the nearest gene, for either wild type or ΔZn cistromes. (D) Heatmap of GR ChIP-Seq coverage, for both wild type specific – and common (wild type and mutant) peaks (n = 2–4). (E) GR ChIP-qPCR for selected loci in MEFs treated with LPS for 3 h and Dex overnight (n = 5), shown as % input together with a negative site. Values are mean ± SEM, ns = not significant, *P< 0.05, **P< 0.01. (F) Western blot of endogenous co-IPs in MEFs treated with LPS for 3 h and with Dex overnight, representative example from n = 3. (G) GR ChIP-qPCR for selected loci in MEFs treated with LPS and Dex for 0–16 h. t-test for each time point versus untreated (0 h). Values are mean ± SEM, ns = not significant, *P< 0.05, **P< 0.01, ***P< 0.001.