Figure 6.
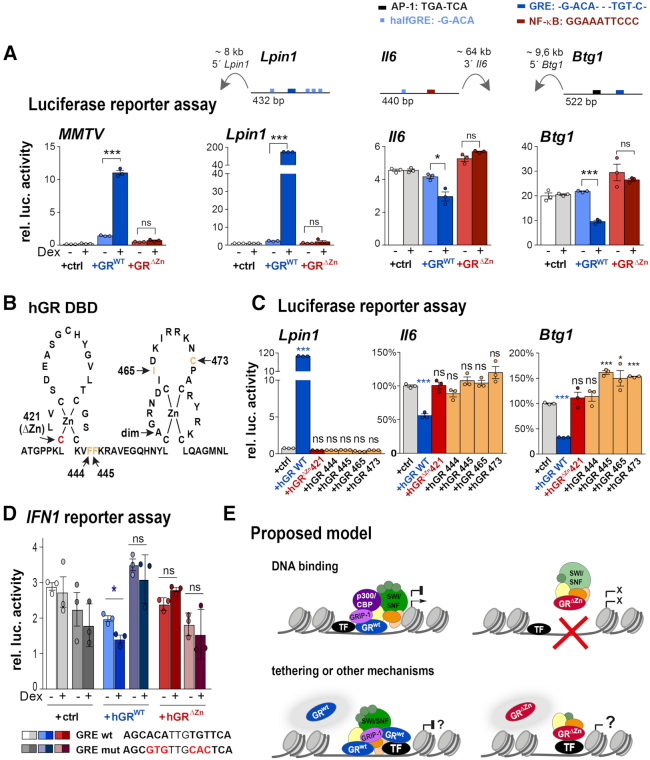
Positive and negative regulation of GR reporters involves DNA binding. (A) Normalized luciferase activity in CV-1 cells overexpressing empty vector (+ctrl), GR wild type or ΔZn mutant and MMTV, Lpin1, Il6 or Btg1 reporters. Cells were treated with LPS and either vehicle or Dex (− or +) overnight (n = 3 biological replicates, values are mean ± SEM, ns = not significant, *P< 0.05, ***P< 0.001). The cartoons depict enhancer positions and composition. (B) Human GR DNA-binding domain (DBD) showing the mutations used in C. (C) Normalized luciferase activity in CV-1 cells overexpressing empty vector (+ctrl), human GR wild type, ΔZn (hGR C421G) and F444G, F445G, I465G and C473G mutants. Cells were treated with vehicle or Dex overnight (n = 3 biological replicates). Values represent mean ± SEM. ns = not significant, *P< 0.05, ***P< 0.001. (D) Normalized luciferase activity in CV-1 cells overexpressing empty vector (+ctrl), human GR wild type or ΔZn (hGR C421G) mutant. Cells were treated with LPS and either vehicle or Dex (− or +) overnight (n = 3 biological replicates, values are mean ± SEM, ns = not significant, *P< 0.05). (E) Proposed mechanism: GR binding to DNA leads to the recruitment of GRIP-1, HATs, the SWI/SNF complex and other co-regulators to activate or repress transcription. The transcriptional inertia of GRΔZn mutants could be explained by the failure to recruit co-regulators, or by other mechanisms such as squelching or non-genomic actions. While tethering still occurs GRΔZn mice, it is not sufficient to regulate gene expression either positively or negatively.