Figure 4.
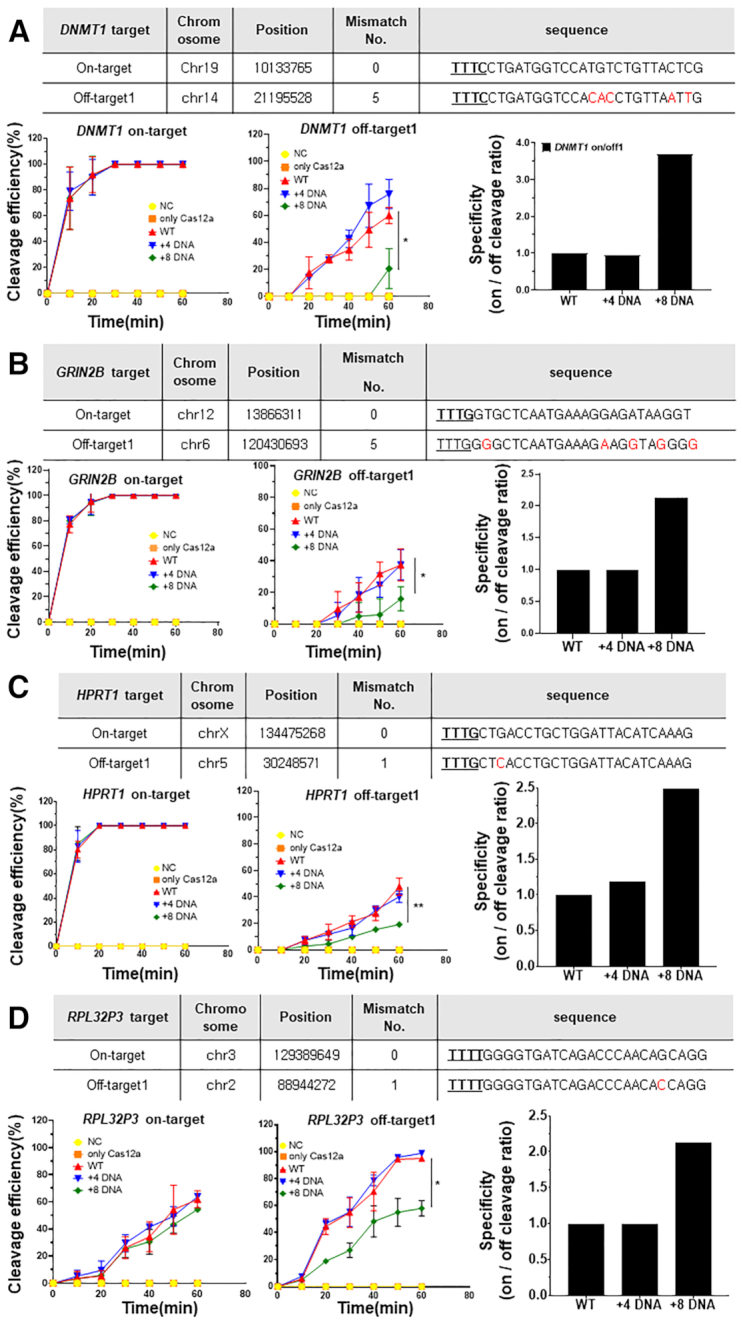
Time-course analysis of the on/off-target cleavage of Cas12a using chimeric DNA–RNA guides on various target sequences. Target and off-target DNA cleavage experiments to confirm the target specificity of AsCpf1 using a chimeric DNA–RNA guide (DNA substitutions in the 3′-end) on (A) DNMT1, (B) GRIN2B, (C) HPRT1 and (D) RPL32P3 genes. Table: Information about on- and off-target sequences, Left: On-target cleavage over time, Middle: Off-target cleavage over time, Right: Target specificity (on/off cleavage ratio (%)) of AsCas12a using chimeric DNA–RNA guides (serial 4-nt and 8-nt DNA substitutions in the 3′-end of the (cr)RNA). NC: negative control, only Cas12a: only protein treated, WT: Wild-type crRNA was treated with Cas12a, +4 DNA: Chimeric crRNA (sequential 4-nt DNA substitution at 3′-end of crRNA) was treated with Cas12a, +8 DNA: Chimeric crRNA (sequential 8-nt DNA substitution at 3′-end of crRNA) was treated with Cas12a. All cleavage efficiencies were calculated from agarose gel separated band intensity (cleaved fragment intensity (%)/total fragment intensity (%)) for 60 min at 10 min interval points and normalized to wild-type (cr)RNA. Data are shown as means ± s.e.m. from three independent experiments. P-values are calculated using a two-tailed Student's t-test (ns: not significant, *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001). On- and off-target sequence information for each gene is shown in table at the top. The PAM sequence is underlined and shown in bold. Mismatch sequences to wild-type reference are shown in red.
