Figure 6.
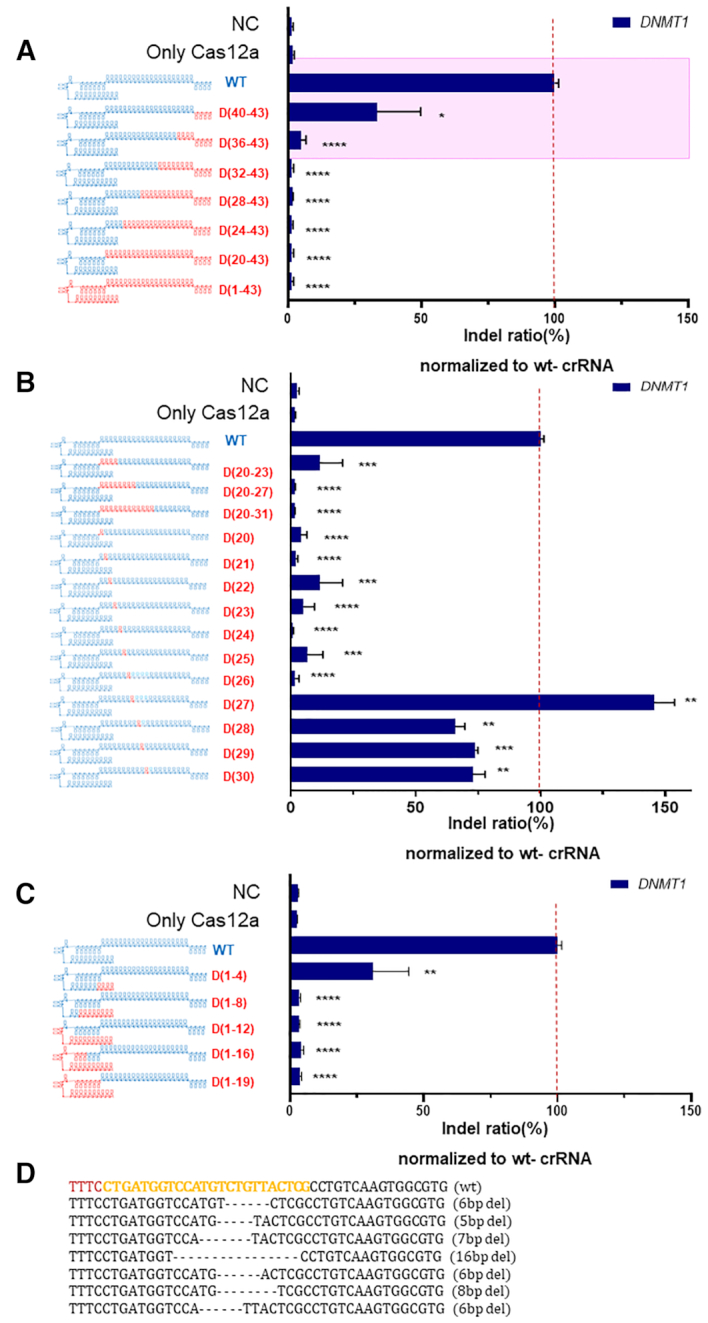
Intracellular genome editing using Cas12a and chimeric DNA–RNA guides. (A) Endogenous locus (DNMT1) editing in HEK293FT cells using chimeric DNA–RNA guides with serial 4-nt DNA substitutions from the 3′-end of the (cr)RNA. The RNA portion of the Cas12a (cr)RNA is shown in blue, and the DNA portion is shown in red ('D' indicates a DNA and the number of substituted DNA nucleotides is indicated). Cas12a genome editing with (cr)RNA with wild-type, 3′-end 4-nt DNA substitution and 3′-end 8-nt substitution is highlighted in the pink box. (B) Endogenous locus (DNMT1) editing in HEK293FT cells using chimeric DNA–RNA guides with DNA substitutions in the seed region of the (cr)RNA. (C) Endogenous locus (DNMT1) editing in HEK293FT cells using chimeric DNA–RNA guides with serial 4-nt DNA substitutions from the 5′-end of the (cr)RNA. (D) Representative genome editing pattern of AsCas12a using chimeric DNA–RNA guides. Sequencing data is obtained from DNMT1 targeted amplicon sequencing (NGS). The deleted base is shown by the dashed line. PAM sequence (TTTN) and target sequence for AsCas12a is shown in brown and yellow color, respectively. All the relative indel ratio was calculated from targeted amplicon sequencing (indel frequency (%) = mutant DNA read number/total DNA read number) and normalized to wild-type (WT) (cr)RNA. Data are shown as means ± s.e.m. from three independent experiments. P-values are calculated using a two-tailed Student's t-test (ns: not significant, *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001).
