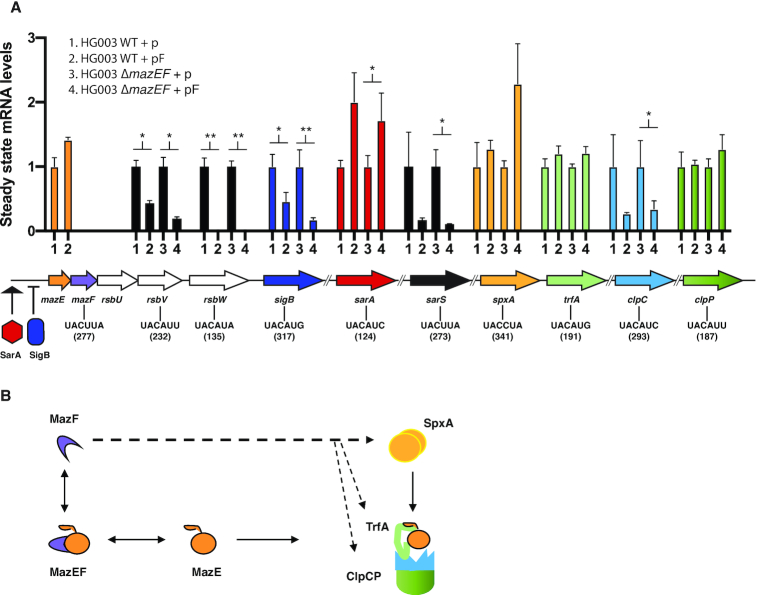
Figure 5.
(A) Steady state transcript levels recorded by qRT-PCR for ten genes in wild-type or mazEF-deleted strains carrying the empty plasmid (p) compared to mazF-plasmid (pF) 10 mpi with ATc. Results are presented as means ± SD of three independent experiments; ** P < 0.01; * P < 0.05 significant differences between the strain carrying the empty plasmid (p) and the strain carrying the mazF-plasmid (pF). The gyrB gene was used to normalize for RNA content. Gene hydrolysis probes hybridize specifically onto the MazF detected site. The MazF recognition sequence and position within the gene is indicated in the gene diagram below. (B) Diagram showing MazF effect on proteins involved in MazE antitoxin proteolysis and containing MazF cleavage motifs. MazF can potentially cleave spxA, trfA, clpC and clpP transcripts (denoted by dashed arrows). SpxA regulates transcription of TrfA adaptor protein, TrfA adaptor in turn is responsible of presenting MazE substrate for degradation to the ClpCP proteolytic system.