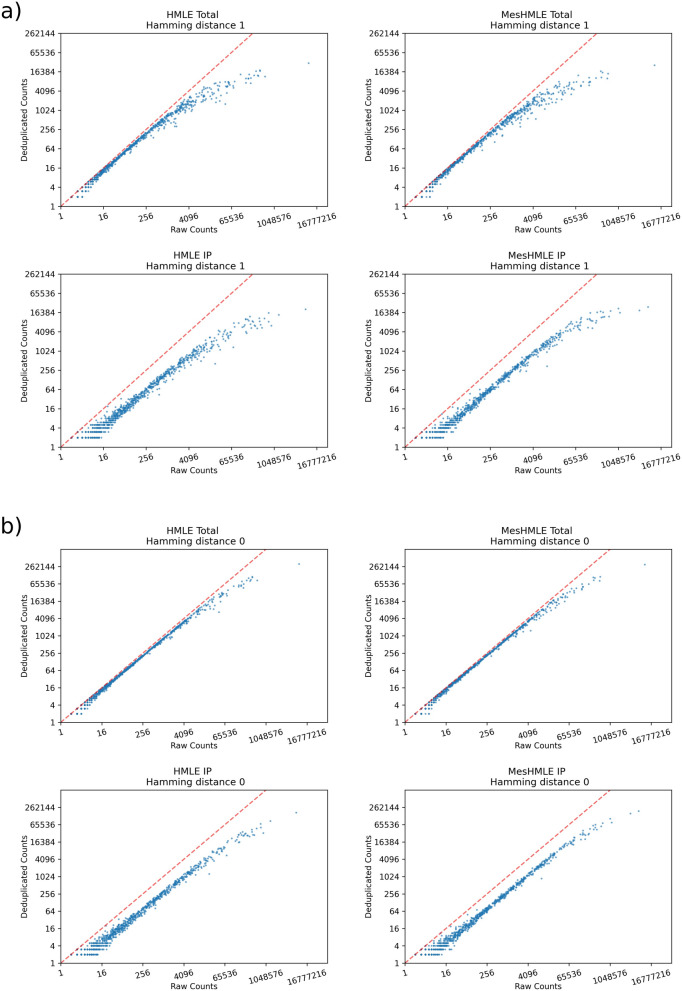
Figure 1.
Highly expressed microRNAs are subject to over de-duplication. 4 separate smRNA libraries (HMLE and MesHMLE cells; total RNA and RNA co-immunoprecipitated with AGO) are shown, with each miRNA represented as dots plotted on axes of total RNA reads (x axis) and de-duplicated read counts (y axis). (a) Draws from a more limited pool of UMIs on account of de-duplicating otherwise identical reads in which there is a single nucleotide mismatch between UMIs (Hamming distance = 1), to account for PCR or sequencing error. In (b) no UMI sequence divergence is accommodated (Hamming distance = 0). For clarity, 4 libraries are represented here. Data from additional biological replicates are included in Supplementary Fig. 1.