Abstract
Breast cancer is a molecularly heterogeneous disease that can be subdivided into different subtypes. Compared with the other subtypes, luminal breast cancer (LBC) is considered more susceptible to bone metastasis. However, the intrinsic mechanisms remain elusive. Bioinformatics analysis of the preset study showed that N-acetyltransferase 1 (NAT1) was specifically expressed in LBC and closely correlated with bone metastasis. In addition, NAT1 could promote LBC cell migration and clonal formation, induce osteoclast differentiation and raise the Rankl/Opg ratio in osteoblasts. Our in vivo experiment demonstrated that NAT1 promoted LBC bone metastasis and bone destruction, which could be reversed by NAT1 inhibitor treatment. The result of cytokine array showed that NAT1 could significantly over activate the NF-κB signaling pathway and up-regulate the expression of IL-1B, which further worked as downstream factors in these processes. All these results demonstrated NAT1 was up-regulated in LBC and promoted the formation of bone metastatic niche and osteolytic bone metastasis through the NAT1/NF-κB/IL-1B axis. This finding may provide a new pathway to help understand the mechanisms of LBC bone metastasis and suggest a novel therapeutic and diagnostic target for its treatment.
Keywords: Luminal breast cancer, bone metastasis, N-acetyltransferase 1 (NAT1), IL-1B, bone microenvironment
Introduction
Breast cancer (BC) is one of the most common malignant tumors in women. There are about one million new cases and 400,000 deaths in the world, and about 270,000 new cases and 70,000 deaths in China every year [1-3]. BC has become the first killer threatening women’s health. Like other malignant tumors, metastasis has become the main reason for the failure of clinical treatment of BC [4]. Once metastasis occurs, it is difficult to control the disease from progressing by the treatments currently available. BC is extremely aggressive, and the bone is one of the most common distant metastatic organs [5]. Bone metastasis usually causes severe pain, pathological fracture, spinal cord compression, hypercalcemia and other skeletal-related events (SREs) [6], which seriously affect the life quality of patients, and bring heavy financial burdens to clinical treatment and home nursing care. The median survival of BC patients with bone metastasis is about 2-3 years, and only 20% patients survive for more than 5 years [7]. Bone metastasis is the “watershed” to accelerate the process of death.
According to the expression patterns of estrogen receptor (ER), progesterone receptor (PR) and growth hormone receptor (Her2) on cancer cells, BC can be subdivided into luminal breast cancer (LBC), Her2 and triple negative (TNBC) subtypes [8]. A large number of studies suggest that different types of BC have different clinical prognoses. LBC is considered a subtype with relatively good prognosis but more susceptible to bone metastases [9-11]. Similar results were also observed in our previous studies. A retrospective study of 87 BC patients with bone metastasis demonstrated LBC cell predilection of bone metastasis [12]. And gene set enrichment analysis (GSEA) in our another previous study showed less aggressive and low-grade features of BC patients with bone metastasis compared with those without bone metastasis, while LBC is more susceptible to bone metastasis [13]. However, the intrinsic mechanisms underlying the regulation of bone metastasis in BC remains elusive.
Transformation from a primary tumor to overt formation of metastasis is a complex process involving the participation of various types of cells [14,15]. Once tumor cells are disseminated into the bone microenvironment, utilization and reconstruction are started and a bone metastatic niche will be built, which disrupts the balance between osteoclasts (OCs) and osteoblasts (OBs) [16-18]. Chemokines and other soluble factors such as interleukin-1B (IL-1B) work as connectors and regulators in this process. IL-1B is a member of the of the IL-1 family as a secreted protein composed of 269 amino acids [19]. The expression of IL-1B is up-regulated in primary tumors and closely related to the clinical prognosis of tumor patients [20,21]. IL-1B secreted by tumor cells is involved in the regulation of tumor bone metastasis and transformation of the bone microenvironment [22].
The main aim of the present study was to screen key molecules that regulate LBC bone metastasis. We found that N-acetyltransferase 1 (NAT1) was upregulated in LBC, suggesting that NAT1 may play a regulatory role in LBC bone metastasis by promoting bone metastatic niche formation. This finding may provide a novel therapeutic and diagnostic target for the management of BC bone metastasis, especially in LBC patients.
Materials and methods
Bioinformatics analysis
BC and normal tissue specimens from the Cancer Genome Atlas (TCGA) were analyzed by expression profile microarrays. Data set GSE2034 and GSE12777 were obtained from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and used for further analysis.
Cells and clinical samples
Human BC cell lines (MCF-10A, HBL-100, Hs578T, BT-549, MDA-MB-231, BT-474, T47D and MCF7) as well as RAW 264.7 and MC3T3-E1 cells were obtained from American Type Culture Collection (ATCC). Bone marrow-derived macrophages (BMMs) and bone marrow-derived stromal cells (BMSCs) were isolated from C57/BL6 mice as previously described [23].
A total of 44 primary BC tissue specimens and 16 BC bone metastatic tissue specimens were obtained from patients who received corresponding surgeries at Changzheng Hospital (Shanghai, China). Informed consent was signed by all the participating patients. Ethical consents were granted by the committees for ethical review of research involving Human Subjects of Navy Medical University (Shanghai, China).
Real-time quantitative PCR (qRT-PCR)
Total RNA was extracted by using TRIzol (Thermo Fisher Scientific, Waltham MA, USA). Reverse transcription and qRT-PCR were performed with PrimeScript™ RT Master Mix and SYBR Premix Ex Taq (Takara, Kyoto, Japan) according to the corresponding instructions. Data were analyzed by 2-ΔΔCt method. Related primers are listed in Table 2.
Table 2.
Primer sequences used in this study
| Gene | Forward primer (5’→3’) | Reverse primer (5’→3’) |
|---|---|---|
| β-actin | GTACGCCAACACAGTGCTG | CGTCATACTCCTGCTTGCTG |
| Human NAT1 | ACTGGCATGATTCACCTT | TTCCCTTCTGATTTGGTC |
| Human IL-1B | CACGATGCACCTGTACGATCA | GTTGCTCCATATCCTGTCCCT |
| Mouse Rankl | ATCCCATCGGGTTCCCATAA | TTCGTGCTCCCTCCTTTCAT |
| Mouse Opg | CAGATGGGTTCTTCTCAGGT | TCTCGGCATTCACTTTGGTC |
Western blot
Total protein was extracted using RIPA buffer. Samples were equally loaded in a 10% sodium dodecyl sulfate gel and transferred to nitrocellulose membranes. After being blocked with 5% bovine serum albumin, the membranes were incubated with the indicated primary antibodies overnight at 4°C, then with DyLight 800-conjugated secondary antibody and scanned with LI-COR Infrared Imaged Odyssey (Gene Company Ltd.). Primary antibodies are as follows: anti-NAT1, anti-IKB-α, anti-p-IKB-α, anti-IKK-β, anti-p-IKK-β, anti-p65, anti-p-p65 (Abways, Shanghai, China), anti-IL-1B (Abcam, UK), anti-Histone H3 (Cell Signaling Technology, USA) and anti-β-actin (Sigma-Aldrich, USA).
Immunohistochemistry (IHC) staining
All the clinical specimens were fixed with 4% paraformaldehyde and embedded in paraffin. Sample processing was based on the standard procedure as previously described [24]. The expression of NAT1 was detected by imaging with a microscope (Leica, Wetzlar, German) and measured with a multiplicative quick score system [25]. The total score of 0-9 was rated as NAT1-low, while the score of 10-18 was rated as NAT1-high.
Construction of NAT1 knockdown and overexpression cell lines
The specific short hairpin RNA (shRNA) plasmids targeting human NAT1 and NAT1 over expression (OE) plasmid were constructed. The shRNA oligos were GTTCCCTTTGAGAACCTTAAC (sh1#) and ATCTACTCCTTTACTCTTAAG (sh2#). The virus was harvested from the supernatant of 293T cells 48-h post-transfection of the shRNA or OE plasmids. T47D and MCF7 cells were further infected with shRNA virus, and BT-549 cells were infected with OE virus.
Cell migration and clonal formation assays
Transwell assays were carried out for cell migration with Boyden chambers (Corning, USA). 5×104-1×105 tumor cells suspended with 100μl serum-free medium were added into the apical chamber, while the basolateral chamber was filled with 600 μl medium supplemented with 2% fetal bovine serum (FBS). After 8-h culture in the cell incubator with T47D and BT-549 cells, and 20 h with MCF7 cells, migrated cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Bright-field images were obtained with an Olympus inverted microscope and migrated cells were counted using Image-Pro Plus 6.0. For clonal formation assay, cells in logarithmic growth were plated into 6-well plates (1000 cells/well) and cultured for at least 7 days. Cells were fixed, stained and imaged thereafter. NAT1 inhibitor and Recombinant Human IL-1B (PeproTech, USA) was added directly in the apical chambers or 6-well plates when needed.
Differentiation of BMMs and BMSCs
Differentiation medium for OC was prepared with 10 ng/ml M-CSF and 50 ng/ml RANKL (R&D, Minneapolis, MN, USA) on the basis of modified α-MEM containing 10% FBS, while the supplements of differentiation medium for OB were 50 µg/ml L-ascorbic acid, 1 mM dexamethasone and 1M β-glycerophosphate. Additionally, conditioned medium (5% for OC differentiation and 10% for OB differentiation) or IL-1B was added into the differentiation medium. For OC differentiation, isolated BMMs were seeded into 96-well plates (1×104 cells/well) and incubated with the OC differentiation medium for 5-7 days. The medium was changed every 2 days. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton-X 100, renatured with 0.1% PBST (Sangon Biotech, Shanghai, China) and stained with tartrate resistant acid phosphatase (TRAP) staining kit (Sigma-Aldrich, USA). For OB differentiation, isolated BMSCs were plated into 24-well plates (1×105 cells/well) and cultured with OB differentiation medium for 7 days. Differentiated cells were subjected to the same process as OC mentioned above, but stained with alkaline phosphatase (ALP) staining kit (Sigma-Aldrich, USA).
Animalmodels
Luciferase-labeled T47D shNC and sh1# cells were inoculated into the left heart ventricles or directly injected into the bone marrow cavity of the right tibia in BALB/c female nude mice aged 4-5 weeks (n= 6 per group). The NAT1 inhibitor (10 μM, 0.1 ml) or PBS was intraperitoneally (i.p.) injected every two days after tumor inoculation into the left heart ventricles to explore the potential treatment effects in BALB/c female nude mice aged 4-5 weeks (n=8 per group). Metastatic conditions were monitored by using an in vivo imaging system (Ivis System, Caliper Life Sciences, USA). Bone metastatic lesions were further radiographically assessed and histopathologically analyzed with H&E and TRAP staining. All mice were randomly divided into two groups and the animal care and experiments were in accordance with the guidelines of the use of laboratory animals of Navy Medical University and approved by the Committee on the Ethics of Animal Experiments of the Navy Medical University.
Luciferase reporter assays
As previously described [26], dual luciferase assays were conducted in a 24-well plate. BT-549 cells were cultured to 70% confluence and transfected with indicated concentrations of NAT1-OE plasmid. After 24-h transfection, firefly and Renilla luciferase levels were quantified sequentially using the Dual Luciferase Assay kit (Promega, USA) following the manufacturer’s recommendations.
Immunofluorescence staining
BT-549 cells were seeded on glass coverslips and transfected with OE-NAT1 or ZsGreen Vector plasmids. After 12-h culture, cells were fixed with 4% paraformaldehyde. After permeabilization and blocking with bovine serum albumin, the cells were incubated with the primary antibody at 4°C overnight and secondary antibody for 2-h at room temperature. The cells were then incubated with DAPI for 5 min and mounted on glass slides for microscopic analysis.
Statistical analysis
The sample size was defined according to our preliminary experiment. In vitro experiments were repeated at least three times in our study. All statistics were analyzed by using SPSS (version 22.0.0, IBM corp., New York, USA) or GraphPad Prism, (version 7, La Jolla, CA, USA). Results are presented as means ± standard deviation (SD). Significant differences were calculated by unpaired Student’s t test, Mann-Whitney U test and one-way ANOVA according to different types of data. Overall survival (OS) and metastasis-free survival (MFS) were measured by the Kaplan-Meier method and analyzed by log-rank tests. P value <0.05 was considered to be statistically significant.
Results
NAT1 is a key regulator in LBC bone metastasis by bioinformatics analysis
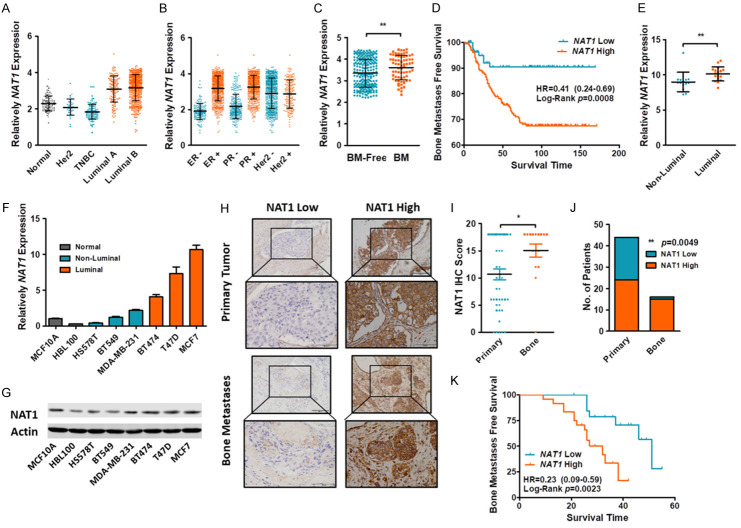
To identify key regulators in in LBC bone metastasis, we first reviewed the TCGA data set which included 726 BC samples and 113 normal controls. Tumor samples were classified into three groups as Her2+, TNBC and LBC. Comparison of different BC subtypes and normal controls identified a series of 58 genes which were specifically expressed in LBC with at least a two-fold change (Figure S1A). As aforementioned, previous studies have reported several gene sets which may play vital roles in regulation of BC bone metastasis. We compared our 58 genes with two published gene sets of bone metastasis (Smid Bone Metastases [27] and Savci Bone Metastases [28]) and finally identified NAT1, which was specifically expressed in both LBC (Figure 1A) and bone metastases gene sets (Figure S1B). Tumor samples were binary divided according to the ER, PR and Her2 status and identified that the expression of NAT1 was closely correlated with ER and PR, but interrelated little with the expression of Her2 (Figure 1B). Therefore, we speculated that NAT1 may work as a key regulator in LBC bone metastasis.
Figure 1.
NAT1 was up-regulated in LBC and correlated with bone metastasis. (A, B) The TCGA database indicate that NAT1 was upregulated in LBC. Meanwhile, the expression of NAT1 was correlated closely with ER and PR. but interrelated little with the expression of Her2. (C) Compared with bone metastasis -free patients. Bone metastatic patients had a higher NAT1 expression in their primary tumors. **P=0.0038 by Mann-Whitney U test. (D) Kaplan-Meier plot of BMFS for patients with primary BCs. nHigh=203. nLow=83. Log-Rank p=0.0008. (E) Expression of NAT1 between Luminal and non-Luminal BC cell lines. **P=0.0018 by Mann-Whitney U test. (F, G) NAT1 expression in normal and BC cell lines. (H-K) IHC staining of primary BC (n=44) and BC bone metastasis (n=16). The expression of NAT1 was significantly up-regulated in bone metastasis (I). *P=0.0193 by unpaired t test. Expression of NAT1 in primary and metastatic BC samples (J). P=0.0049 by one-way ANOVA. Kaplan-Meier plot of primary BC tumors in bone metastasis-free patients (K). nHigh=24. nLow=20. Log-Rank P=0.0023.
The dataset GSE2034 [29] consisting of 286 primary BC patients was used for further confirmation. NAT1 was significantly up-regulated in ER+ BC patients, which are mainly LBCs. The expression of NAT1 was closely correlated with the LBC markers (Figure S2). NAT1 expression in the primary tumors of patients with bone metastasis was higher than that in those with no bone metastasis (Figure 1C). Also, patients with high NAT1 expression in their primary BC tumors were more susceptible to bone metastasis but not to brain or lung metastasis (Figures 1D and S3). Comparison of BC cell lines also revealed significantly high expression of NAT1 in the Luminal groups (Figure 1E).
Expression of NAT1 in cell lines and bone metastasis samples
The expression of NAT1 was detected in two normal breast and six BC cell lines (3 TNBCs and 3 LBCs) by qRT-PCR and Western blot, and the result showed that NAT1 was highly expressed in LBC (Figure 1F, 1G). Clinically, 44 primary BC and 16 bone metastasis specimens were subjected to IHC staining. As expected, the expression of NAT1 was significantly up-regulated in the bone metastasis specimens (15/16, 93.75%) (Figure 1H, 1I), and only in 54.55% (24/44) of the primary BC specimens. Primary BC patients were further followed up to observe bone MFS. Survival curves indicated that patients with high NAT1 expression were more susceptible to bone metastasis.
NAT1 promotes LBC cell migration and clonal formation, and induces the bone metastatic niche
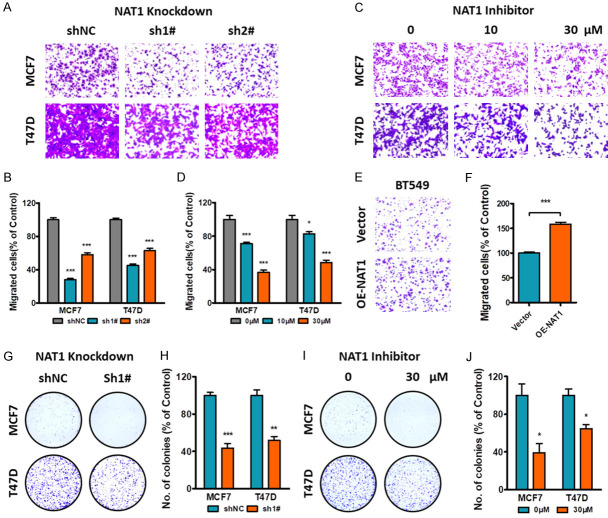
The expression of NAT1 was stably down-regulated in two LBC cell lines T47D and MCF7 by shRNAs, and stably over-expressed in the TNBC cell line BT-549 (Figure S4). Transwell assay revealed that depletion of NAT1 by either shRNAs or specific inhibitors could significantly reduce cell migration. As expected, NAT1 overexpression promoted BT-549 cell migration (Figure 2A-F). Meanwhile, clonal formation was also attenuated either by shNAT1 or inhibitor treatment on LBC cell lines (Figure 2G-J).
Figure 2.
Effect of NAT1 on BC migration and clone formation. A, B. Knockdown of NAT1 in LBC cell lines reduced cell migration by transwell assay. ***P<0.001 by unpaired t test. C, D. Cell migration was depressed by NAT1 inhibitor at indicated concentrations. *P<0.05. ***P<0.001 by unpaired t test. E, F. Upregulation of NAT1 expression in TNBC cell line BT-549 promoted tumor migration. ***P<0.001 by unpaired t test. G-J. NAT1 knockdown or NAT1 inhibitor significantly depressed tumor clonal formation in LBC cell lines. *P<0.05. ***P<0.001 by unpaired t test.
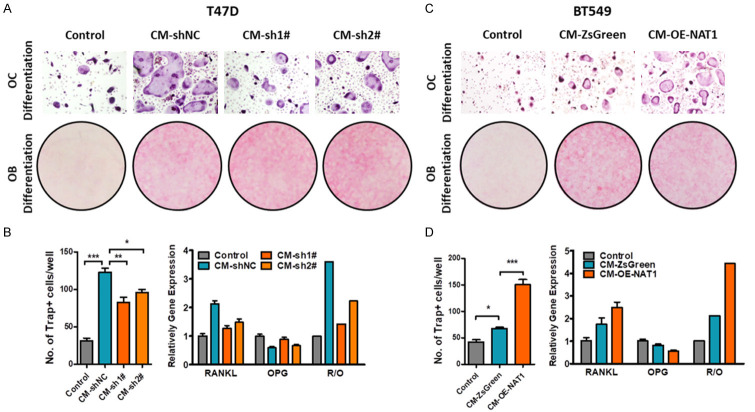
The Published literature indicates that tumor cells can induce an organ specific niche for metastasis formation and progression. Such findings are also applied to bone metastasis. To assess the effect of NAT1 on bone metastatic niche, tumor conditioned media were collected and added to the culture systems of OC and OB differentiation for simulation of the BC-induced bone metastasis niche. The results showed that the role of the tumor conditioned media in promoting osteoclastogenesis was attenuated when NAT1 was knocked down (Figure 3A up and 3B left). However, tumor conditioned media harvested from BT-549 cells with NAT1 overexpression promoted the process of OC differentiation (Figure 3C up and 3D left).
Figure 3.
Effect of NAT1 expression on tumor induced bone metastatic niche. (A, B) Knockdown of NAT1 attenuated LBC-promoted bone metastatic niche. Tumor conditioned media were collected and added to the culture systems of OC and OB differentiation to simulate the BC-induced bone metastatic niche. Knockdown of NAT1 significantly decreased tumor induced OC differentiation (A up and B left. One-way ANOVA. *P<0.05. **P<0.01. ***P<0.001) but had little effect on OB differentiation (A down). However. knockdown of NAT1 significantly decreased tumor induced Rankl expression in OBs and upregulated Opg expression. One-way ANOVA. pRankl=0.0003. pOpg=0.0027. (C, D) Upregulation of NAT1 in BT-549 cells promoted tumor induced OC differentiation (C up and D left. One-way ANOVA. *P<0.05. ***P<0.001), and increased Rankl expression but decreased Opg expression in OBs. One-way ANOVA. pRankl=0.0111. pOpg=0.0050.
With respect to OBs, the tumor conditioned media could not only promote cell differentiation, but regulate the expression of Rankl and Opg. These results indicate that NAT1 had a weak effect on OB differentiation (Figure 3A down and 3C down), but significantly affected the process of gene expression (Figure 3B right and 3D right). NAT1 Knockdown in tumor cells down-regulated the expression of Rankl and up-regulated Opg, and decreased the Rankl/Opg ratio. Reversely, NAT1 overexpression promoted Rankl and depressed Opg, eventually increasing the Rankl/Opg ratio.
NAT1 promotes LBC osteolytic bone metastasis in vivo
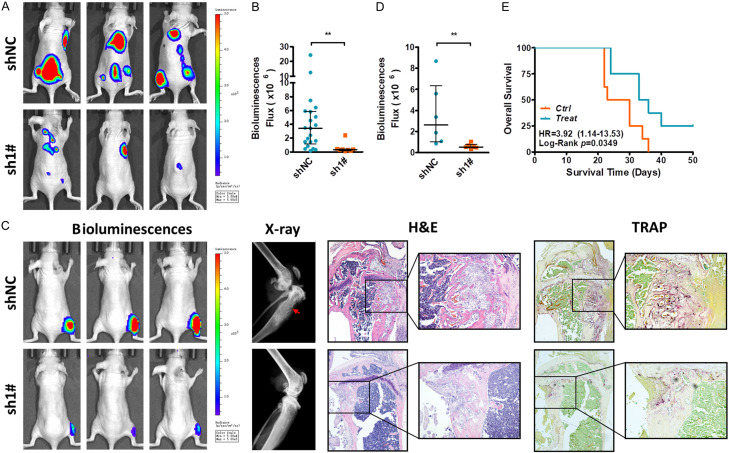
To further confirm the effect of NAT1 on LBC bone metastasis, three animal models were used for in vivo investigation. Firstly, luciferase labeled T47D-shNC and sh1# cells were inoculated into the left heart ventricle of the nude mice and cultured for 3 weeks. Metastasis involvement was measured in vivo and the mice were sacrificed thereafter. Lesions detected by in vivo images (Figure 4A, 4B) and further confirmed by autopsy (Table 1). In mice administered with shNC cells, tumor metastasis was detected in all 6 mice and 16 of the 21 lesions involving the bone structures. However, metastasis was detected in 4 of the 6 mice with sh1# cells and only 3 involving bone structures of the total 8 lesions. These results suggested that knockdown of NAT1 significantly hindered LBC metastasis, especially to the bone.
Figure 4.
Inhibition of NAT1 attenuates LBC osteolytic bone metastasis. A, B. Luciferase labeled T47D-shNC and sh1# cells were inoculated into the left heart ventricle of the nude mice and cultured (n=6 for each group). Mice were sacrificed and metastasis was evaluated after 3 weeks. **P=0.0016 by Mann-Whitney U test. C, D. Luciferase labeled T47D-shNC and sh1# cells were directly injected in to the right tibia bone marrow cavity of the nude mice (n=6 per group). After two-week culture, the mice were sacrificed and local bone destruction was detected by X-ray and Trap staining. **P=0.0043 by Mann-Whitney U test. E. After successful inoculation of T47D cells into the left heart ventricles. 16 nude mice were equally randomized into, and treated either with the NAT1 inhibitor (10 uM. 0.1 ml) or PBS every two days by i.p. injection. Mice were cultured till death or to 50 days after tumor inoculation. Survival time was recorded and Kaplan-Meier plot was drawn accordingly. Log-Rank P=0.0349.
Table 1.
Tumor metastases of the left heart ventricle model
| Group | No. of mice with metastases | No. of lesions | No. of Bone lesions | Bone/Total |
|---|---|---|---|---|
| shNC | 6/6 | 21 | 16 | 76.2% |
| sh1# | 4/6 | 8 | 3 | 37.5% |
The second model was used to investigate the effect of NAT1 on tumor induced bone metastatic niche. Luciferase labeled T47D-shNC and sh1# cells were directly injected in to the right tibia bone marrow cavity of nude mice (n=6 for each group). In vivo luciferase signals were detected after two weeks and indicated advanced lesions in shNC groups (Figure 4C, 4D). Extensive bone destruction and overt OC activation were detected by X-ray scan and Trap staining after mice were sacrificed (Figure 4C).
Finally, we investigated the potential therapeutic effect of the NAT1 inhibitor. After inoculation of the tumor into the left heart ventricles, mice were treated with the NAT1 inhibitor (10 uM, 0.1 ml) or PBS every two days by i.p. After a 50-day follow-up period, all mice in control group died with a median survival of 26.5 days and 6 of the 8 mice in the treatment group died with a median survival of 34.5 days (Figure 4E). All these results strongly indicate the vital roles of NAT1 in regulating LBC osteolytic bone metastasis.
IL-1B is regulated by NAT1 through NF-κB signaling pathway
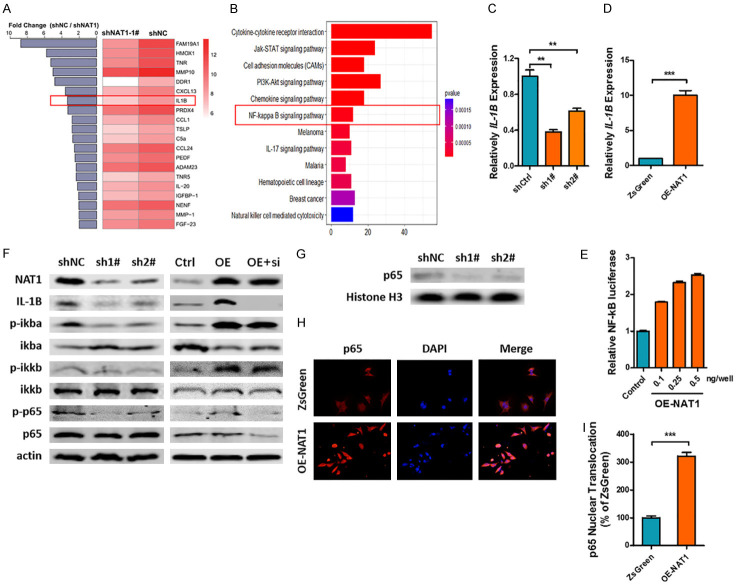
Our previous finding indicated that NAT1 could regulate the bone metastasis niche by tumor conditioned media. As NAT1 is an intracellular enzyme, there should be secretable downstream cytokines that act as regulators in the process. Hence, we collected the tumor conditioned media and subjected them to cytokine array analysis. A total of 267 differentially expressed genes (DEGs) were detected and further subjected to GO and KEGG analyses. The results indicated that the downstream cytokines played vital roles in cell migration, motility and chemotaxis (Figure S5). Altogether 140 proteins were significantly down-regulated in sh1# cells and the top 20 are listed in Figure 5A. Special attention was paid to IL-1B, a key regulator of tumor progression and metastasis in various types of cancers. Meanwhile, KEGG analysis also revealed that the NF-κB signaling pathway, a key regulation pathway in IL-1B expression, was significantly inhibited after NAT1 depletion (Figure 5B).
Figure 5.
NAT1 promotes IL-1B expression through NF-κB signaling pathway. A, B. After 12-h culture, the conditioned media of T47D-shNAT and sh1# cells were subjected to cytokine array analysis. IL-1B was found to be significantly downregulated after NAT1 knockdown while the KEGG analysis indicated the inhibition of NF-κB signaling pathway. C, D. qRT-PCR confirmed the regulatory effect of NAT1 on IL-1B expression. **P<0.01. ***P<0.001 by unpaired t test. E. Relative NF-κB luciferase activity of BT-549 cells after transfection with OE-NAT1 vectors. One-way ANOVA. P<0.001. F, G. Western blot array indicated that NAT1 significantly affected the phosphorylation of NF-κB signaling pathway and p65 nuclear transportation. H, I. Immunofluorescence indicated that NF-κB p65 nuclear transportation was significantly upregulated after OE-NAT1 transfection in BT-549 cells. ***P<0.001 by unpaired t test.
To confirm the finding by cytokine array, we first performed the qRT-PCT and demonstrated that NAT1 played a role in regulating the expression of IL-1B at mRNA level (Figure 5C, 5D). Secondly, a luciferase reporter gene assay revealed that the NF-κB luciferase activity of BT-549 cells was significantly upregulated after OE-NAT1 vectors transfection in a dose-dependent manner (Figure 5E). Western blot indicated that NAT1 knockdown inhibited the phosphorylation of ikba, ikkb, and NF-κB p65, which ultimately depressed the expression of IL-1B. And as expected, overt activation of the NF-κB signaling pathway and upregulation of IL-1B expression were observed after transfection of the OE-NAT1 vector into TNBC BT-549 cells. However, co transfection of siRNAs targeting NF-κB p65 (sip65) together with the OE-NAT1 vector abolished the upregulation of IL-1B expression (Figure 5F). Finally, nuclear p65 detection and immunofluorescence images further confirmed the regulatory effect of NAT1 on NF-κB signaling pathway (Figure 5G-I).
NAT1/NF-κB/IL-1B axis promotes LBC osteolytic bone metastasis
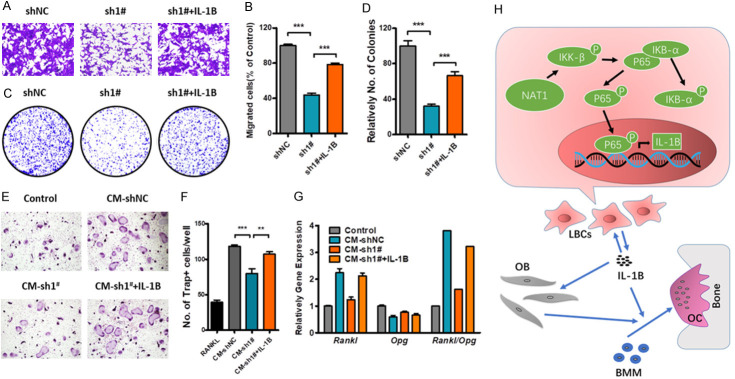
Several rescue experiments were performed to confirm the role of IL-1B in regulating LBC bone metastasis and bone destruction. As shown in Figure 6, IL-1B could rescue the processes of cell migration, clone formation and tumor induced OC differentiation. Meanwhile, IL-1B could also restore the Rankl/Opg ratio in OBs. Taken together, our findings demonstrated that NAT1 was up-regulated in LBCs, and promoted the formation of bone metastatic niche and osteolytic bone metastases through the NAT1/NF-κB/IL-1B axis (Figure 6H).
Figure 6.
IL-1B promotes LBC osteolytic bone metastasis. A-G. IL-1B could rescue NAT1 depletion induced downregulation of BC migration. clone formation. OC differentiation and OB Rankl/Opg expression. One-way ANOVA. **P<0.01. ***P<0.001. H. A schematic illustration of the NAT1/NF-κB/IL-1B axis in regulation of the LBC bone metastatic niche and osteolytic bone metastasis.
Discussion
Many studies have demonstrated that LBC is more susceptible to bone metastasis than other types of BC. Smid et al. analyzed 334 cases of primary BC and found that 48 (67.6%) of the 71 bone metastases were associated with LBC [10]. Zhang et al. summarized 715 cases of various types of BCs and divided them into two groups according to the expression of ER. The results showed that bone metastasis was more likely to occur in LBC patients with positive ER [30]. Foulkes et al. reviewed more than 600 related studies and found that the lung metastasis rate of patients with TNBC was high as 40%, with a bone metastasis rate of 10% vs. 40% in non-TNBC patients [11]. Sihto et al. focused on the cumulative organ distribution of tumor metastasis and found that there were 321 metastatic lesions in 234 cases, of which the highest incidence was bone metastasis (n=127, 39.6%), and most of them were from LBC [9]. Based on these clinical findings, we speculated that genes specifically expressed in LBC may contribute to the intrinsic mechanism in regulating bone metastasis. A total of 58 genes were identified and compared with published bone metastases gene expression signatures thereafter. Finally, NAT1 was screened as the candidate.
NAT1 is an enzyme involved in the biotransformation of many aromatic and heterocyclic amines [31]. A large number of related studies have shown that NAT1 and its gene polymorphism are closely related to tumor development [32,33]. The expression of NAT1 was significantly increased in ER + breast cancer and closely related to the degree of malignancy [34,35]. Tiang et al. downregulated the expression of NAT1 by shRNA in three negative BC cell lines, and found that the migration and metastasis abilities of BC tumor cells were decreased significantly [36]. In the current study, we first demonstrated that the expression of NAT1 was closely related to ER expression and consistent with luminal markers GATA3 and FOXA1. Our clinical data also confirmed the interrelationship between NAT1 expressions and bone metastasis.
After colonization, disseminated tumor cells will reconstruct the bone microenvironment by interacting with bone marrow stromal cells, which disrupts the balance of bone metabolization and finally constructs a bone metastatic niche [7,15,37]. BCs are featured with mixed patterns of both excessive bone destruction and reactive bone formation, which requires the participation of both OCs and OBs [38,39]. Our results indicated that NAT1 overexpression could not only directly promote OC differentiation but also restore the Rankl/Opg ratio in OBs.
As NAT1 is an intracellular enzyme, there should be secretable downstream cytokines that act as regulators in the process. Our cytokine array finally identified IL-1B as a potential target of NAT1. IL-1B is an inflammatory chemokine which plays important roles in tumor initiation and metastasis [40,41]. Primary BC patients with high IL-1B expression were found to be more susceptible to bone metastasis [42]. IL-1B expression was up regulated in bone tropism subclones compared with parental MDA-MB-231 cells [43]. Liu et al. reported that bone metastasis was significantly reduced when IL-1B was knocked out, while IL-1B overexpression increased prostate cancer bone colonization [44]. Compared with those in the bone microenvironment, IL-1B secreted by tumor cells play more important roles in metastasis and bone implantation [22]. Tumor-derived IL-1B can promote OB proliferation and cytokine secretion by the latter cells, which can help bone colonization [22,45]. Meanwhile, the osteolytic ability of OCs induced by IL-1B was significantly enhanced under pathological conditions [46]. Therefore, inhibition of IL-1B signaling pathway can not only inhibit tumor growth, but target the bone microenvironment, inhibit osteoclast differentiation, reduce the release of bone-derived growth factors, and control tumor bone metastasis [47]. It was found in our study that NAT1 could promote IL-1B expression via the NF-κB signaling pathway, and IL-1B could not only promote BC cell proliferation and colonization but help construct the bone metastatic niche. The NAT1/NF-κB/IL-1B axis works as an intrinsic mechanism in LBC bone metastasis.
Previous studies suggested that low-level NAT1 was associated with tamoxifen resistance and that NAT1 could be a candidate marker to predict the effect of antiestrogen therapy, which reveals the potential regulatory function of ER inhibitor on NAT1 [48,49]. Moreover, it can be learned from several studies that endocrine therapy is able to inhibit IL-1B mRNA levels and that IL-1B could influence tamoxifen resistance through the methylation of ERα gene [50,51]. These conclusions may remind us of the possibility that endocrine therapy could play a regulatory role in LBC bone metastasis via the NAT1/NF-κB/IL-1B axis. However, bone metastasis is a complex process with multiple mechanisms, the insights of which remain to be further explored.
In summary, our study provided a new pathway may help understand the mechanism underlying LBC bone metastasis. NAT1 was found to be upregulated in LBC, which was closely associated with tumor bone metastasis by promoting tumor cell migration and colonization, bone metastasis niche formation and osteolytic bone metastasis through the NAT1/NF-κB/IL-1B axis.
Acknowledgements
This work was supported by key project funding in the basic research field of the Shanghai Municipal Science and Technology Commission (17JC1400903) and the National Natural Science Foundation of China (81871470, 81902732, 81972505).
Disclosure of conflict of interest
None.
Abbreviations
- NAT1
N-acetyltransferase 1
- BC
Breast cancer
- SREs
skeletal-related events
- ER
estrogen receptor
- PR
progesterone receptor
- Her2
growth hormone receptor 2
- LBC
Luminal breast cancer
- TNBC
triple negative breast cancer
- GSEA
gene set enrichment analysis
- OC
osteoclast
- OB
osteoblast
- IL-1B
Interleukin-1B
- NC
Negative control
- OE
over expression
- M-CSF
macrophage colony-stimulating factor
- RANKL
The receptor activator of nuclear factor-κB ligand
- TRAP
tartrate resistant acid phosphatase
- ALP
alkaline phosphatase
- Opg
Osteoprotegerin
- H&E
Hematoxylin-eosin
- OS
overall survival
- MFS
metastasis-free survival
- IKB-α
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor alpha
- IKK-β
inhibitor of nuclear factor kappa-B kinase subunit beta
- p65
nuclear factor NF-kappa-B p65 subunit
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China. 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Warner E. Clinical practice. Breast-cancer screening. N Engl J Med. 2011;365:1025–1032. doi: 10.1056/NEJMcp1101540. [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Kang Y. Emerging therapeutic targets in metastatic progression: a focus on breast cancer. Pharmacol Ther. 2016;161:79–96. doi: 10.1016/j.pharmthera.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai WL, Huang WD, Li B, Chen TR, Li ZX, Zhao CL, Li HY, Wu YM, Yan WJ, Xiao JR. microRNA-124 inhibits bone metastasis of breast cancer by repressing Interleukin-11. Mol Cancer. 2018;17:9. doi: 10.1186/s12943-017-0746-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Zhu W, Biskup E, Yang W, Yang Z, Wang H, Qiu X, Zhang C, Hu G, Hu G. Incidence, risk factors and prognostic characteristics of bone metastases and skeletal-related events (SREs) in breast cancer patients: a systematic review of the real world data. J Bone Oncol. 2018;11:38–50. doi: 10.1016/j.jbo.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jinnah AH, Zacks BC, Gwam CU, Kerr BA. Emerging and established models of bone metastasis. Cancers (Basel) 2018;10:176. doi: 10.3390/cancers10060176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 9.Sihto H, Lundin J, Lundin M, Lehtimäki T, Ristimäki A, Holli K, Sailas L, Kataja V, Turpeenniemi-Hujanen T, Isola J, Heikkilä P, Joensuu H. Breast cancer biological subtypes and protein expression predict for the preferential distant metastasis sites: a nationwide cohort study. Breast Cancer Res. 2011;13:R87. doi: 10.1186/bcr2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, Foekens JA, Martens JW. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68:3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 11.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 12.Zhao C, Zhang Z, Zhong N, Fan T, Gao X, Wu Z, Li Z, Liu T, Xiao J. Outcomes and prognostic factors for surgically treated patients with breast cancer spine metastases. J Bone Oncol. 2018;12:38–43. doi: 10.1016/j.jbo.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao C, Lou Y, Wang Y, Wang D, Tang L, Gao X, Zhang K, Xu W, Liu T, Xiao J. A gene expression signature-based nomogram model in prediction of breast cancer bone metastases. Cancer Med. 2019;8:200–208. doi: 10.1002/cam4.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obenauf AC, Massagué J. Surviving at a distance: organ-specific metastasis. Trends Cancer. 2015;1:76–91. doi: 10.1016/j.trecan.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W, Bado I, Wang H, Lo HC, Zhang XH. Bone metastasis: find your niche and fit in. Trends Cancer. 2019;5:95–110. doi: 10.1016/j.trecan.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown HK, Ottewell PD, Evans CA, Holen I. Location matters: osteoblast and osteoclast distribution is modified by the presence and proximity to breast cancer cells in vivo. Clin Exp Metastasis. 2012;29:927–938. doi: 10.1007/s10585-012-9481-5. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Yu C, Gao X, Welte T, Muscarella AM, Tian L, Zhao H, Zhao Z, Du S, Tao J, Lee B, Westbrook TF, Wong ST, Jin X, Rosen JM, Osborne CK, Zhang XH. The osteogenic niche promotes early-stage bone colonization of disseminated breast cancer cells. Cancer Cell. 2015;27:193–210. doi: 10.1016/j.ccell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fili S, Karalaki M, Schaller B. Mechanism of bone metastasis: the role of osteoprotegerin and of the host-tissue microenvironment-related survival factors. Cancer Lett. 2009;283:10–19. doi: 10.1016/j.canlet.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39:1003–1018. doi: 10.1016/j.immuni.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ridker PM, MacFadyen JG, Thuren T, Everett BM, Libby P, Glynn RJ. Effect of interleukin-1β inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet. 2017;390:1833–1842. doi: 10.1016/S0140-6736(17)32247-X. [DOI] [PubMed] [Google Scholar]
- 21.Lewis AM, Varghese S, Xu H, Alexander HR. Interleukin-1 and cancer progression: the emerging role of interleukin-1 receptor antagonist as a novel therapeutic agent in cancer treatment. J Transl Med. 2006;4:48. doi: 10.1186/1479-5876-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tulotta C, Lefley DV, Freeman K, Gregory WM, Hanby AM, Heath PR, Nutter F, Wilkinson JM, Spicer-Hadlington AR, Liu X, Bradbury SMJ, Hambley L, Cookson V, Allocca G, Kruithof de Julio M, Coleman RE, Brown JE, Holen I, Ottewell PD. Endogenous production of IL-1B by breast cancer cells drives metastasis and colonisation of the bone microenvironment. Clin Cancer Res. 2019;25:2769–2782. doi: 10.1158/1078-0432.CCR-18-2202. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Huang J, Wang F, Li W, Wu X, Zhao C, Zhao J, Wei H, Wu Z, Qian M, Sun P, He L, Jin Y, Tang J, Qiu W, Siwko S, Liu M, Luo J, Xiao J. Dual targeting of bile acid receptor-1 (TGR5) and farnesoid X receptor (FXR) prevents estrogen-dependent bone loss in mice. J Bone Miner Res. 2019;34:765–776. doi: 10.1002/jbmr.3652. [DOI] [PubMed] [Google Scholar]
- 24.Wang D, Zhao C, Gao L, Wang Y, Gao X, Tang L, Zhang K, Li Z, Han J, Xiao J. NPNT promotes early-stage bone metastases in breast cancer by regulation of the osteogenic niche. J Bone Oncol. 2018;13:91–96. doi: 10.1016/j.jbo.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Gu Y, Han Y, Zhang Q, Jiang Z, Zhang X, Huang B, Xu X, Zheng J, Cao X. Tumor exosomal RNAs promote lung pre-metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell. 2016;30:243–256. doi: 10.1016/j.ccell.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 26.Zhao C, Wang D, Tang L, Zhang Z, Li S, Qian M, Wu Z, Zhou W, Liu M, Luo J, Liu T, Li Z, Xiao J. Stromal cell-derived CCL20 promotes tumor progression and osteolysis in giant cell tumor of bone. Cell Physiol Biochem. 2018;51:2472–2483. doi: 10.1159/000495903. [DOI] [PubMed] [Google Scholar]
- 27.Smid M, Wang Y, Klijn JG, Sieuwerts AM, Zhang Y, Atkins D, Martens JW, Foekens JA. Genes associated with breast cancer metastatic to bone. J. Clin. Oncol. 2006;24:2261–2267. doi: 10.1200/JCO.2005.03.8802. [DOI] [PubMed] [Google Scholar]
- 28.Savci-Heijink CD, Halfwerk H, Koster J, van de Vijver MJ. A novel gene expression signature for bone metastasis in breast carcinomas. Breast Cancer Res Treat. 2016;156:249–259. doi: 10.1007/s10549-016-3741-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 30.Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, Foekens JA, Massagué J. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues-Lima F, Dairou J, Busi F, Dupret JM. Human arylamine N-acetyltransferase 1: a drug-metabolizing enzyme and a drug target. Curr Drug Targets. 2010;11:759–766. doi: 10.2174/138945010791170905. [DOI] [PubMed] [Google Scholar]
- 32.Laurieri N, Crawford MH, Kawamura A, Westwood IM, Robinson J, Fletcher AM, Davies SG, Sim E, Russell AJ. Small molecule colorimetric probes for specific detection of human arylamine N-acetyltransferase 1, a potential breast cancer biomarker. J Am Chem Soc. 2010;132:3238–3239. doi: 10.1021/ja909165u. [DOI] [PubMed] [Google Scholar]
- 33.Butcher NJ, Minchin RF. Arylamine N-acetyltransferase 1: a novel drug target in cancer development. Pharmacol Rev. 2012;64:147–165. doi: 10.1124/pr.110.004275. [DOI] [PubMed] [Google Scholar]
- 34.Tozlu S, Girault I, Vacher S, Vendrell J, Andrieu C, Spyratos F, Cohen P, Lidereau R, Bieche I. Identification of novel genes that co-cluster with estrogen receptor alpha in breast tumor biopsy specimens, using a large-scale real-time reverse transcription-PCR approach. Endocr Relat Cancer. 2006;13:1109–1120. doi: 10.1677/erc.1.01120. [DOI] [PubMed] [Google Scholar]
- 35.Wakefield L, Robinson J, Long H, Ibbitt JC, Cooke S, Hurst HC, Sim E. Arylamine N-acetyltransferase 1 expression in breast cancer cell lines: a potential marker in estrogen receptor-positive tumors. Genes Chromosomes Cancer. 2008;47:118–126. doi: 10.1002/gcc.20512. [DOI] [PubMed] [Google Scholar]
- 36.Tiang JM, Butcher NJ, Minchin RF. Effects of human arylamine N-acetyltransferase I knockdown in triple-negative breast cancer cell lines. Cancer Med. 2015;4:565–574. doi: 10.1002/cam4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurizi A, Rucci N. The osteoclast in bone metastasis: player and target. Cancers (Basel) 2018;10:218. doi: 10.3390/cancers10070218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shupp AB, Kolb AD, Mukhopadhyay D, Bussard KM. Cancer metastases to bone: concepts, mechanisms, and interactions with bone osteoblasts. Cancers (Basel) 2018;10:182. doi: 10.3390/cancers10060182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mantovani A, Barajon I, Garlanda C. IL-1 and IL-1 regulatory pathways in cancer progression and therapy. Immunol Rev. 2018;281:57–61. doi: 10.1111/imr.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steel JL, Terhorst L, Collins KP, Geller DA, Vodovotz Y, Kim J, Krane A, Antoni M, Marsh JW, Burke LE, Butterfield LH, Penedo FJ, Buysse DJ, Tsung A. Prospective analyses of cytokine mediation of sleep and survival in the context of advanced cancer. Psychosom Med. 2018;80:483–491. doi: 10.1097/PSY.0000000000000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coleman R, Cameron D, Dodwell D, Bell R, Wilson C, Rathbone E, Keane M, Gil M, Burkinshaw R, Grieve R, Barrett-Lee P, Ritchie D, Liversedge V, Hinsley S, Marshall H. Adjuvant zoledronic acid in patients with early breast cancer: final efficacy analysis of the AZURE (BIG 01/04) randomised open-label phase 3 trial. Lancet Oncol. 2014;15:997–1006. doi: 10.1016/S1470-2045(14)70302-X. [DOI] [PubMed] [Google Scholar]
- 43.Nutter F, Holen I, Brown HK, Cross SS, Evans CA, Walker M, Coleman RE, Westbrook JA, Selby PJ, Brown JE, Ottewell PD. Different molecular profiles are associated with breast cancer cell homing compared with colonisation of bone: evidence using a novel bone-seeking cell line. Endocr Relat Cancer. 2014;21:327–341. doi: 10.1530/ERC-13-0158. [DOI] [PubMed] [Google Scholar]
- 44.Liu Q, Russell MR, Shahriari K, Jernigan DL, Lioni MI, Garcia FU, Fatatis A. Interleukin-1beta promotes skeletal colonization and progression of metastatic prostate cancer cells with neuroendocrine features. Cancer Res. 2013;73:3297–3305. doi: 10.1158/0008-5472.CAN-12-3970. [DOI] [PubMed] [Google Scholar]
- 45.Templeton ZS, Lie WR, Wang W, Rosenberg-Hasson Y, Alluri RV, Tamaresis JS, Bachmann MH, Lee K, Maloney WJ, Contag CH, King BL. Breast cancer cell colonization of the human bone marrow adipose tissue niche. Neoplasia. 2015;17:849–861. doi: 10.1016/j.neo.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiratori T, Kyumoto-Nakamura Y, Kukita A, Uehara N, Zhang J, Koda K, Kamiya M, Badawy T, Tomoda E, Xu X, Yamaza T, Urano Y, Koyano K, Kukita T. IL-1beta induces pathologically activated osteoclasts bearing extremely high levels of resorbing activity: a possible pathological subpopulation of osteoclasts, accompanied by suppressed expression of kindlin-3 and talin-1. J Immunol. 2018;200:218–228. doi: 10.4049/jimmunol.1602035. [DOI] [PubMed] [Google Scholar]
- 47.Holen I, Lefley DV, Francis SE, Rennicks S, Bradbury S, Coleman RE, Ottewell P. IL-1 drives breast cancer growth and bone metastasis in vivo. Oncotarget. 2016;7:75571–75584. doi: 10.18632/oncotarget.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bièche I, Girault I, Urbain E, Tozlu S, Lidereau R. Relationship between intratumoral expression of genes coding for xenobiotic-metabolizing enzymes and benefit from adjuvant tamoxifen in estrogen receptor alpha-positive postmenopausal breast carcinoma. Breast Cancer Res. 2004;6:R252–63. doi: 10.1186/bcr784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SJ, Kang HS, Jung SY, Min SY, Lee S, Kim SW, Kwon Y, Lee KS, Shin KH, Ro J. Methylation patterns of genes coding for drug-metabolizing enzymes in tamoxifen-resistant breast cancer tissues. J Mol Med (Berl) 2010;88:1123–31. doi: 10.1007/s00109-010-0652-z. [DOI] [PubMed] [Google Scholar]
- 50.Rodenas MC, Cabas I, García-Alcázar A, Meseguer J, Mulero V, García-Ayala A. Selective estrogen receptor modulators differentially alter the immune response of gilthead seabream juveniles. Fish Shellfish Immunol. 2016;52:189–97. doi: 10.1016/j.fsi.2016.03.041. [DOI] [PubMed] [Google Scholar]
- 51.Jiménez-Garduño AM, Mendoza-Rodríguez MG, Urrutia-Cabrera D, Domínguez-Robles MC, Pérez-Yépez EA, Ayala-Sumuano JT, Meza I. IL-1β induced methylation of the estrogen receptor ERα gene correlates with EMT and chemoresistance in breast cancer cells. Biochem Biophys Res Commun. 2017;490:780–785. doi: 10.1016/j.bbrc.2017.06.117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.