Abstract
Cyclosporine (CsA) is characterized by a narrow therapeutic window and high interindividual pharmacokinetic variability, particularly in juvenile patients. The aims of this study were to build a population pharmacokinetic model of CsA in Chinese children with hematopathy who received allogeneic hematopoietic stem cell transplantation (allo-HSCT) and to identify covariates affecting CsA pharmacokinetics. A total of 86 Chinese children aged 8.4 ± 3.8 years (range 1.1–16.8 years) who received allo-HSCT were enrolled. Whole blood samples were collected before allo-HSCT. Genotyping was performed using an Agena MassARRAY system. A total of 1010 trough plasma concentration values of CsA and clinical data were collected. The population pharmacokinetic model of CsA was constructed using nonlinear mixed-effects modeling (NONMEM) software. The stability and performance of the final model were validated using bootstrapping and normalized prediction distribution errors. We showed that a one-compartment model with first-order elimination adequately described the pharmacokinetics of CsA. The typical values for clearance (CL) and volume of distribution (V) were 42.3 L/h and 3100 L, respectively. Body weight, postoperative days, CYP3A4*1 G genotype, estimated glomerular filtration rate and coadministration of triazole antifungal drugs were identified as significant covariates for CL. Weight and postoperative days were significant covariates for the V of CsA. Our model can be adopted to optimize the CsA dosing regimen for Chinese children with hematopathy receiving allo-HSCT.
Keywords: cyclosporine, population pharmacokinetic model, hematopoietic stem cell transplantation, Chinese children
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT) has become an effective treatment option for refractory or recurrent hematological malignancies such as leukemia, lymphoma and myelodysplastic syndrome [1, 2]. However, graft-versus-host disease (GVHD) is one of the main reasons for the failure of transplantation. Cyclosporine (CsA), a calcineurin inhibitor, has been widely used to prevent GVHD disease in patients who have undergone allo-HSCT [3]. Indeed, CsA is characterized by a narrow therapeutic window and high interindividual pharmacokinetic variability. Thus, therapeutic drug monitoring (TDM) is advised to adjust doses and reduce toxicity, especially in juvenile patients, who exhibit a wider range of pharmacokinetic variability than adults [4]. However, owing to the hysteresis of TDM, identification of the factors affecting the pharmacokinetics of CsA is essential for improving safety and efficacy.
Population pharmacokinetics (Pop-PK), which can identify and quantify the influences of different factors on drug exposure, is a powerful and potential tool for individualized dosing [5]. Although Pop-PK studies of CsA have been extensively performed in recent years, only two have been undertaken in juvenile allo-HSCT patients [6, 7]. According to reports, genetic polymorphisms have important implications for CsA pharmacokinetics [8–10]. CsA is mainly metabolized by the cytochrome P450 (CYP) isoenzymes CYP3A4 and CYP3A5 (mainly in the liver, and to a lesser extent in the kidney and intestine) [11, 12] and is also a substrate of the P-glycoprotein (encoded by the ABCB1 gene), which is involved in the absorption and elimination of CsA [13, 14]. Polymorphisms in the genes of those proteins could influence the pharmacokinetics of CsA [8, 15–17]. Because cytochrome P450 oxidoreductase (POR) is the only obligate electron donor for all hepatic microsomal CYP enzymes [18], POR genetic polymorphisms could also affect CsA pharmacokinetics [10, 19]. In addition, the pregnane X receptor (PXR) has been identified as the key nuclear receptor regulating the expression of CYP3A4, CYP3A5, and ABCB1 [20, 21]. Furthermore, several studies have also examined the effect of other genetic polymorphisms in ABCC2 [20, 22, 23], PPARA [19], RELA [24], NFKBIA [24], TNFAIP3 [25], FOXP3 [26] and CYP2C19 [22, 27] on CsA pharmacokinetics. However, gene polymorphism factors were not included in the existing Pop-PK studies in juvenile allo-HSCT patients conducted by Sarem et al. [6] or Ni et al. [7]. Therefore, a thorough understanding of the effects of genetic polymorphisms is essential for individualized cyclosporine therapy.
The aims of the present study were to establish a population pharmacokinetic model of CsA in children receiving allo-HSCT considering gene polymorphism and to identify the covariates influencing CsA pharmacokinetics.
Materials and methods
Patients and data collection
A total of 1010 drug monitoring data points were collected retrospectively from clinical reports of 86 child recipients who were first administered CsA and then underwent allo-HSCT because of malignant hematologic disorders at Peking University People’s Hospital, Beijing, China.
Data on the following characteristics were collected from electronic medical records: age, sex, body weight (WT), height, body mass index (BMI), hemoglobin (HB), hematocrit (HCT), albumin (Alb), serum creatinine (Scr), total bilirubin (T-BIL), alanine transaminase (ALT), aspartate transaminase (AST), estimated glomerular filtration rate (eGFR), postoperative day (POD), type of disease (TOD), and concomitant medications. This study was approved by the Health Authority Ethics Committee of the Peking University People’s Hospital and conducted in accordance with the Declaration of Helsinki. All patients signed an informed consent approved by Ethics Committee.
CsA administration
Patients received intravenous CsA (Sandimmun, liquid solution, Neoral, Novartis Pharma, Basel, Switzerland) beginning on Day −7 or −10 of transplantation as an intermittent intravenous infusion over 2 h every 12 h with an initial dose of 2–3 mg/kg. The daily dosage was then adjusted according to trough concentration (C0) to achieve a target concentration of 150–250 ng/mL. The administration route was changed to oral when patients were able to tolerate oral intake after transplantation.
CsA sampling and concentration assay
All blood samples were drawn before intravenous CsA infusion in the morning, and the concentrations were trough concentrations. CsA concentrations were detected using a chemiluminescent microparticle immunoassay (CMIA) that was performed on the Architect R i2000SR platform (Abbott, Chicago, IL, USA). This detection method has a lower limit of quantitation of 30 ng/mL and a linearity ranging from 30 to 1500 ng/mL, with a coefficient variation of less than 4%.
DNA extraction and genotyping
Genomic DNA was extracted from whole blood samples with a genomic DNA purification kit (Promega, Madison, WI, USA). The genomic DNA was stored at −20 °C until use. Single nucleotide polymorphism (SNP) genotyping analysis for CYP3A5*3 (rs776746), CYP3A5 (rs15524), CYP3A4*1 G (rs2242480), CYP3A4 (rs4646437), CYP3A4*18B (rs28371759), ABCB1 c.1236 C > T (rs1128503), ABCB1 c.2677 G > T (rs2032582), ABCB1 c.3435 C > T (rs1045642), POR*28 (rs1057868), POR (rs17685), POR (rs3823884), POR (rs1135612), PXR C-25385T (rs3814055), PXRG24113A (rs2276706), PXR (rs6785049), PXR (rs3842689), PXR (rs1523127), ABCC2 c.3972 C > T (rs3740066), PPARA (rs4823613 A > G), RELA (rs7119750), RELA (rs11820062), NFKBIA G2758A (rs696), TNFAIP3 (rs2230926), FOXP3 (rs3761549), CYP2C19*17 (rs12248560), CYP2C19*2 (rs4244285) and CYP2C19*3 (rs4986893) was carried out using an Agena MassARRAY system (Agena Bioscience, USA). Duplicate samples and negative controls were included to ensure accuracy in genotyping.
Pharmacokinetic analysis
Pharmacokinetic analysis was performed using the nonlinear mixed-effects modeling program NONMEM VII (V2.0; Icon Development Solutions, USA). Pharmacokinetic parameters and associated variability were estimated with the first-order conditional estimation (FOCE) method with the interaction.
Structural model
The one-compartment model with first-order elimination was tested. Interindividual variability of the pharmacokinetic parameters was estimated using an exponential model expressed as follows: (Eq. 1), while the residual error using a mixed model expressed as follows: (Eq. 2).
| 1 |
where Pi is the predicted value of the parameter for the i-th patient, Ppop is the typical population value of the pharmacokinetic parameter, and ηi is a random variable, identically distributed with a mean of 0 and variance of ω2, that was used to describe the variation for the individual i.
| 2 |
where Cobs is the observed concentration and Cpred is the predicted concentration. ε1 and ε2 are presumed to obey the distribution of N (0, σ12) and N (0, σ22), respectively.
Covariate analysis
Based on the nonlinear relationship between body weight and drug elimination capacity in children, pharmacokinetic parameters were modified by the allometric scaling theory, which is expressed as follows:
| 3 |
where Pi and WTi are the pharmacokinetic parameter and body weight of the i-th individual, respectively. Pstd is the parameter of an individual with a standard weight of 70 kg. PWR is the allometric coefficient fixed at a value of 0.75 for clearance and a value of 1 for distribution volume [7, 28–30].
The covariates screened in this study included the following variables: age, WT, height, BMI, HB, HCT, ALT, AST, Alb, Scr, T-BIL, eGFR, POD, sex, concomitant medications, TOD, and genetic polymorphism.
A forward inclusion and backward elimination process was used to select covariates. The objective function value (OFV) provided by NONMEM was used as a diagnostic criterion. A decrease in OFV greater than 3.84 after introducing a certain covariate indicated that the covariate had a significant effect on the fitting of the model (P < 0.05). The contribution of each covariate was verified by a stepwise backward elimination (P < 0.01, the OFV value increased by more than 6.63).
Model validation
The quality of the Pop-PK model was extensively evaluated using goodness-of-fit (GOF) plots including observations vs. individual predictions, observations vs. population predictions, conditional weighted residuals vs. population predictions and conditional weighted residuals vs. time.
The bootstrap method was used to evaluate the stability of the final model. During this process, partial data from the original datasets were resampled 500 times. The parameters of each subgroup dataset were calculated. Stability was assessed based on the frequency distribution of these parameters.
The normalized prediction distribution error (NPDE) method was also used to validate the final model [31]. NPDE is appropriate for the design of multiple-dose drug administration with limited sampling points. In this study, a total of 1000 simulations were constructed to generate the corresponding NPDE values for validation of the final model.
Results
Demographics
Eighty-six subjects (56 boys and 30 girls) were recruited for this population pharmacokinetic study. The mean ± SD age was 8.38 ± 3.78 years (range 1.1–16.8), the mean ± SD body weight was 31.93 ± 16.75 kg (range 6.5–69.0), and the mean ± SD height was 130.28 ± 25.2 cm (range 77–181). The main characteristics are shown in Table 1. The frequency distributions of the CYP3A4, CYP3A5, ABCB1, POR, PXR, ABCC2, PPARA, RELA, NFKBIA, TNFAIP3, FOXP3 and CYP2C19 polymorphisms are presented in Table 2. As shown in Table 2, the minimum allele frequencies of these SNPs in the Chinese population were very close to those recorded in the National Center for Biotechnology Information.
Table 1.
Baseline characteristics of 86 children
| Number | Mean | SD | Median | Range | |
|---|---|---|---|---|---|
| Number of patients | 86 | ||||
| Number of observation | 1010 | ||||
| Dose (mg) | 28.6 | 14.01 | 25 | 5–125 | |
| Gender (male/female) | 56/30 | ||||
| Age (year) | 8.38 | 3.78 | 8.35 | 1.1–16.8 | |
| Weight (kg) | 31.93 | 16.75 | 28.8 | 9.4–78.5 | |
| Height (cm) | 130.28 | 25.2 | 129 | 77–181 | |
| BMI | 17.5 | 4.2 | 16.1 | 12.6–44.1 | |
| Type of disease, n (%) | |||||
| AA | 23 | 26.7% | |||
| ALL | 29 | 33.7% | |||
| AML | 22 | 25.6% | |||
| NHL | 5 | 5.8% | |||
| MDS | 4 | 4.7% | |||
| Other | 3 | 3.5% | |||
| POD | 29.7 | 11.7 | 25 | 0–67 | |
| Laboratory test values | |||||
| HB, g/L | 85.49 | 14.63 | 83.35 | 3.1–811 | |
| HCT, % | 24.42 | 2.88 | 24.26 | 22.−95.4 | |
| ALT, U/L | 48.39 | 33.20 | 40.25 | 4–890 | |
| AST, U/L | 36.42 | 18.54 | 31.75 | 2.9–414 | |
| ALB, U/L | 34.98 | 4.68 | 34.83 | 15–371 | |
| Scr, μmoL/L | 27.40 | 12.84 | 25.5 | 4–215 | |
| T-BIL, μmoL/L | 10.11 | 7.21 | 8.58 | 1.6–205.2 | |
| eGFR, mL/min/1.73 m2 | 179.4 | 34.86 | 172.46 | 24.93–377.54 | |
| Co-medication | |||||
| Triazole | |||||
| None | 4 | ||||
| Voriconazole | 61 | ||||
| Itraconazole | 19 | ||||
| Fluconazole | 1 | ||||
| Posaconazole | 6 | ||||
| Phenytoin | 82 | ||||
| Phenobarbital | 49 | ||||
| Calcium channel blocker | |||||
| None | 51 | ||||
| Nifedipine | 32 | ||||
| Amlodipine | 9 | ||||
AA aplastic anemia, ALL acute lymphocytic leukemia, AML acute myeloid leukemia, NHL non-Hodgkin’s lymphoma, MDS myelodysplastic syndrome
Table 2.
Frequency distributions of gene polymorphisms in the current
| Gene | Variants | n (%) | MAFa (%) | MAFb (%) | |
|---|---|---|---|---|---|
| CYP3A4*1 G | CC | 50 (61.0) | C | 23.2 | 24.4 |
| TT | 6 (7.3) | ||||
| CT | 26 (31.7) | ||||
| CYP3A4*18B | GG | 0 (0) | G | 1.9 | 1.8 |
| AA | 76 (96.2) | ||||
| AG | 3 (3.8) | ||||
| CYP3A4 rs4646437 | GG | 62 (77.5) | A | 13.8 | 13.4 |
| AA | 4 (5.0) | ||||
| GA | 14 (17.5) | ||||
| CYP3A5 rs15524 | GG | 6 (7.3) | G | 29.9 | 28.0 |
| AA | 39 (47.6) | ||||
| AG | 37 (45.1) | ||||
| CYP3A5*3 | CC | 42 (51.9) | T | 27.2 | 28.0 |
| TT | 5 (6.2) | ||||
| CT | 34 (42) | ||||
| ABCB1 c. 3435 C > T | GG | 29 (31.5) | A | 39.7 | 38.7 |
| AA | 10 (10.9) | ||||
| GA | 53 (57.6) | ||||
| ABCB1 c. 1236 C > T | GG | 7 (8.5) | G | 37.2 | 29.3 |
| AA | 28 (34.1) | ||||
| AG | 47 (57.3) | ||||
| ABCB1 c. 2677 G > T | AA | 15 (18.3) | G | 43.9 | 43.9 |
| CC | 16 (19.5) | ||||
| TT | 1 (1.2) | ||||
| AT | 10 (12.2) | ||||
| CA | 32 (39.0) | ||||
| CT | 8 (9.8) | ||||
| POR rs1135612 | GG | 18 (19.6) | G | 42.9 | 42.0 |
| AA | 31 (33.7) | ||||
| GA | 43 (46.7) | ||||
| POR*28 | CC | 37 (45.1) | T | 35.4 | 36.8# |
| TT | 13 (15.9) | ||||
| TC | 32 (39) | ||||
| POR rs17685 | GG | 36 (45) | A | 35.0 | 28.0 |
| AA | 12 (15) | ||||
| GA | 32 (40) | ||||
| POR rs3823884 | GG | 1 (1.3) | A | 21.1 | 25.0 |
| TT | 45 (59.2) | ||||
| GT | 30 (39.5) | ||||
| PXR rs6785049 | GG | 28 (34.1) | A | 44.5 | 45.3 |
| AA | 19 (23.2) | ||||
| AG | 35 (42.7) | ||||
| PXR rs3842689 | DEL.GAGAAG | 31 (37.8) | DEL | 25.0 | NA |
| GAGAAG | 46 (56.1) | ||||
| DEL | 5 (6.1) | ||||
| PXR C-25385T | CC | 47 (57.3) | T | 24.4 | 23.3 |
| TT | 5 (6.1) | ||||
| CT | 30 (36.6) | ||||
| PXR G24113A | GG | 41 (53.2) | A | 29.2 | 20.9 |
| AA | 9 (11.7) | ||||
| GA | 27 (35.1) | ||||
| PXR rs1523127 | CC | 5 (6.1) | C | 25.0 | 23.8 |
| AA | 46 (56.1) | ||||
| CA | 31 (37.8) | ||||
| ABCC2 c.3972 C > T | CC | 51 (63.0) | T | 19.8 | 17.0 |
| TT | 2 (2.5) | ||||
| CT | 28 (34.6) | ||||
| PPARA rs4823613 A > G | GG | 7 (8.5) | G | 28.0 | 23.3 |
| AA | 43 (52.4) | ||||
| AG | 32 (39) | ||||
| RELA rs7119750 | CC | 31 (37.8) | T | 41.4 | 38.9 |
| TT | 17 (20.7) | ||||
| TC | 34 (41.5) | ||||
| RELA rs11820062 | CC | 31 (37.8) | T | 39.0 | 37.2 |
| TT | 13 (15.9) | ||||
| CT | 38 (46.3) | ||||
| NFKBIA G2758A | CC | 30 (36.6) | T | 38.4 | 37.2 |
| TT | 11 (13.4) | ||||
| TC | 41 (50) | ||||
| TNFAIP3 | GG | 0 (0) | G | 3.1 | 7.3 |
| TT | 76 (93.8) | ||||
| GT | 5 (6.2) | ||||
| FOXP3 | CC | 65 (80.2) | T | 14.8 | 13.4 |
| TT | 8 (9.9) | ||||
| TC | 8 (9.9) | ||||
| CYP2C19*2 | GG | 43 (52.4) | A | 28.0 | 25.6 |
| AA | 7 (8.5) | ||||
| GA | 32 (39) | ||||
| CYP2C19*3 | GG | 72 (87.8) | A | 7.3 | 5.8 |
| AA | 2 (2.4) | ||||
| AG | 8 (9.8) | ||||
| CYP2C19*17 | CC | 80 (97.6) | T | 2.4 | 2.2 |
| TT | 0 (0) | ||||
| CT | 2 (2.4) |
MAF minimum allele frequency, NA not available
aresult in present study
bresult recorded in NCBI
#MAF in Asian
Population pharmacokinetic model analysis
A total of 1010 trough blood CsA concentration values were included in this population pharmacokinetic study (a mean of 12 samples per child). The sampling time of TDM was mostly distributed during the first 8–10 h after the last administration, with a narrow distribution range.
A one-compartment model with mixed error adequately described the PK profile. The final model was identified: 5 covariates, including WT, POD, CYP3A4*1 G, eGFR and coadministration of triazole antifungals (TAF), were observed to significantly affect the CL, and 2 covariates, including WT and POD, significantly affected the V of CsA. Detailed information for the remaining parameter estimations is shown in Eq. 4, Eq. 5 and Table 3. The proportional error was 22.9%, and the additive error was 30.3 ng/mL, which was relatively small. The relative standard error (RSE) of all the parameters was lower than 30%, suggesting that the parameters were estimated with good precision and that they gave reliable results.
| 4 |
| 5 |
(If the gene is 1, CLi = CLi*0.984; If the gene is 2 or 3, CLi = CLi*1.22 in Eq. 4, patients were coadministered of TAF, the TAF = 1, otherwise TAF = 0.)
Table 3.
Population pharmacokinetic parameters of CsA and bootstrap results (n = 500)
| Parameter | Final model Estimate (RSE%) | Final model CI (95%) | Bootstrap Median | Bootstrap CI (95%) |
|---|---|---|---|---|
| CL (L/h) | 42.3 (10.6) | 33.48, 51.12 | 42.16 | 32.56, 51.94 |
| V (L) | 3100 (13.1) | 2306.2, 3893.8 | 3018.87 | 2340.67, 4008.99 |
| θTAF-CL | 0.36(16.3) | 0.245, 0.475 | 0.35 | 0.226, 0.459 |
| θPOD-CL | 0.00703 (21.5) | 0.004, 0.01 | 0.0072 | 0.0044, 0.0107 |
| θPOD-V | 0.0197 (29.8) | 0.008, 0.031 | 0.0199 | 0.0084, 0.0351 |
| θeGFR-CL | 0.545 (29.9) | 0.226, 0.864 | 0.542 | 0.204, 0.932 |
| Gene-1 ON CL | 0.984 (8.1) | 0.827, 1.141 | 0.991 | 0.823, 1.189 |
| Gene-2 ON CL | 1.22 (10.1) | 0.979, 1.461 | 1.223 | 0.947, 1.536 |
| Gene-3 ON CL | 1.22 (8.9) | 1.008, 1.432 | 1.221 | 0.986, 1.458 |
| Inter-individual variability | ||||
| ω2 CL | 0.0744 (19.5) | Shrinkage-8.4% | 0.0705 | 0.044, 0.101 |
| ω2 V | 0.454 (24) | Shrinkage-27.2% | 0.447 | 0.249, 0.660 |
| Residual variability | ||||
| σ21 (%) | 0.154 (22.9) | 0.085, 0.223 | 0.152 | 0.081, 0.231 |
| σ22 (ng/mL) | 30.3 (21.7) | 17.442, 43.158 | 29.8 | 16.81, 42.86 |
CL clearance, V distribution volume, Gene = CYP3A4*G (1 for CYP3A4*G CC, 2 for CYP3A4*G TT, 3 for CYP3A4*G CT), TAF triazole antifungals, RSE% relative standard error, CI confidence interval
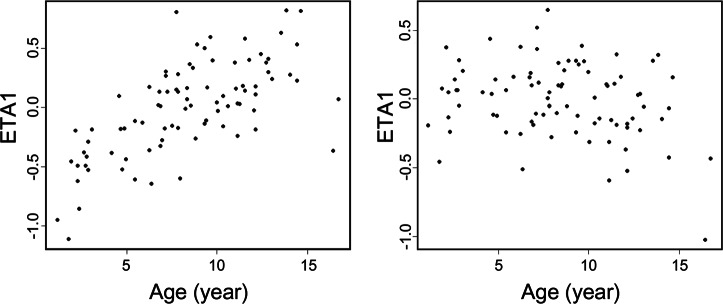
The weight-normalized clearance was 0.75 L·h−1·kg−1. Before and after allometric weight normalization, the comparison of the effect of age on the distribution of the model parameter clearance rate is illustrated in Fig. 1. After incorporation of body weight with allometric scaling, the effect of age on the eta of clearance rate disappeared.
Fig. 1.
Clearance distribution (before and after normalization to allometric weight) plotted against age
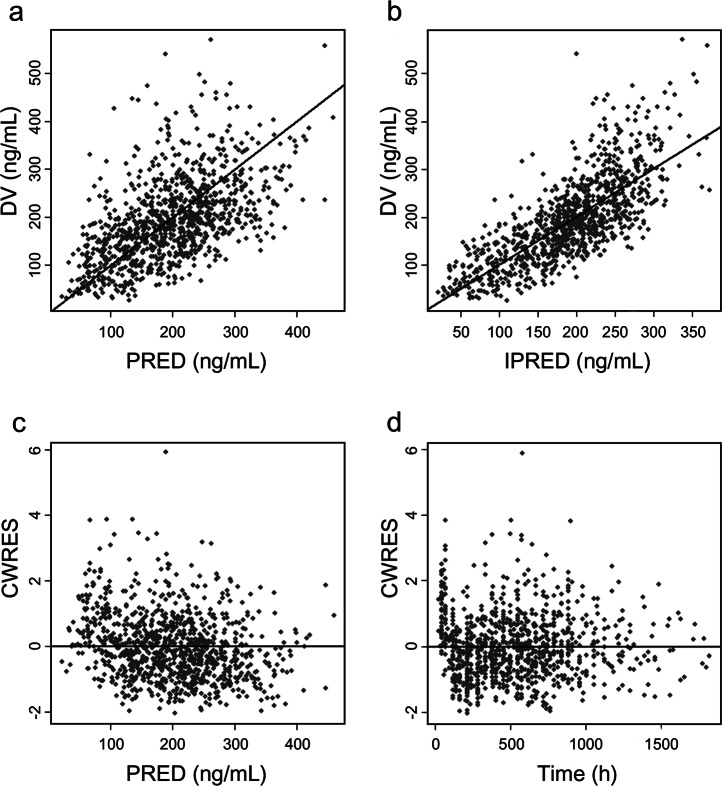
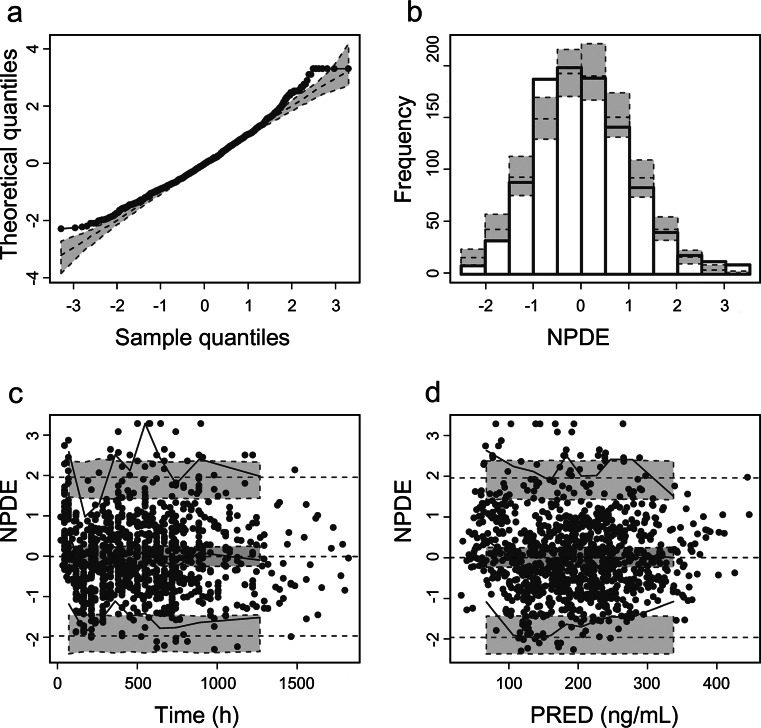
The GOF plots for the final model are shown in Fig. 2. The success rate for the sampling of 500 bootstraps was 97.8%, and the estimated values of the parameters were similar to the median values resulting from the bootstrap method. In addition, the 95% CI contained the typical estimates from the final model and had a narrow range, indicating that the results of the model parameters were reliable and that the model was relatively stable (Table 3). Based on the NPDE results, most of the points in the Q-Q diagram are distributed around the Y = X line (Fig. 3). In addition, the average value on the bar distribution graph was close to 0, and the standard deviation was close to 1, showing a near-normal distribution. These results indicate that the model accurately described the dataset.
Fig. 2.
Diagnostic goodness-of-fit plots. a DV vs. PRED; b DV vs. IPRED; c CWRES vs. PRED; d CWRES vs. time. DV, observed concentrations; PRED population predicted concentrations, IPRED individual predicted concentrations, CWRES conditional weighted residuals
Fig. 3.
Normalized prediction distribution error (NPDE) analysis of the final model. a Q-Q plot of the distribution of the NPDE vs. the theoretical N (0, 1) distribution; b histogram of the distribution of the NPDE; c NPDE vs. time after the first dose, d NPDE vs. PRED
Discussion
This study is the first to describe the population pharmacokinetics of cyclosporine in Chinese children with malignant hematologic disorders who received allo-HSCT. The factors that have been reported to have a potential impact on cyclosporine metabolism were evaluated comprehensively and included developmental, biological and clinical factors as well as gene polymorphisms. Our results showed that a one-compartment model incorporating allometric scaling with first-order elimination was adequate for data modeling. Moreover, body weight, POD, CYP3A4*1 G, eGFR and coadministration of TAF had significant impacts on CL, while body weight and POD had significant impacts on V.
Currently, a weight-based dosing regimen is the most common method of administering cyclosporine in patients receiving hematopoietic stem cell transplants, which was confirmed by our study. In this study, allometrically scaled body weight was shown to be significantly associated with both CL and V, which is consistent with many previous studies [7, 32, 33]. However, the effect of age on CL disappeared after incorporating body weight into the model. This result was consistent with previous results in pediatric renal transplant patients [7, 33]. The probable reason is that body weight and age are closely related to children’s growth and development.
Although genetic polymorphisms may have an effect on CsA pharmacokinetics, few studies have evaluated the effects of genetic polymorphisms on the population pharmacokinetics of CsA in allo-HSCT patients [22, 34]. Our Pop-PK research comprehensively evaluated genetic polymorphisms as covariates in juvenile allo-HSCT patients, which showed that the clearance rate of CsA in CYP3A4*1 G T allele carriers increased by 24.5% compared with that in CYP3A4*1 G CC carriers. However, we found that CYP3A5 and ABCB1 polymorphisms were not significant covariates in the Pop-PK model of CsA. Dai et al. [35] reported that the in vitro intrinsic metabolic clearance estimated from total metabolite formation is approximately 2.3-fold greater for CYP3A4 than for CYP3A5. Both Dai’s and our results suggested that CYP3A4 could play a more dominant role than CYP3A5 in the metabolism of CsA. Thus far, the effects of CYP3A5 and ABCB1 polymorphisms on the pharmacokinetics of CsA are still contradictory, although many studies have been performed [9, 16, 36, 37]. Differences in study population, sample size, ethnicities, concomitant medication and especially the postoperative day might be possible confounding factors.
Children receiving immunosuppressive therapy often take TAF, including voriconazole, itraconazole, fluconazole, and posaconazole, to treat and prevent fungal infections. Consistent with many previous studies [34, 38, 39], our results showed that coadministration of TAF, a class of potent inhibitors of cytochrome P450 3A enzymes [40, 41], was significantly associated with lower CL of CsA in Chinese children who received allo-HSCT. However, none of these medicines were identified as having effects on CsA pharmacokinetics by Zhou et al. [42]. An insufficient number of patients for each drug combination may be the cause of this result.
The known CsA PK changed with the course of postoperative day. There were several conflicting studies about the influence of POD on CL or V. Both Irtan et al. [33] and Okada et al. [43] reported a statistically significant increase in CL/F with POD after renal transplantation. In contrast, Wu et al. suggested a decrease in CL/F via an increase in bioavailability with POD in kidney transplantation patients [44]. Jacobson et al. also reported similar findings after hematopoietic stem cell transplantation [45]. However, these studies all involved oral CsA administration, and no final consensus has yet been reached. Our results showed that the CL of CsA in Chinese juvenile allo-HSCT patients during the first 9 d after transplantation increased and then began to decrease significantly. The underlying cause may be that a high dose of steroids, CYP3A inhibitors, were used during the early stages before transplantation, and the dose was tapered down slowly. Thus, the increase in CL in the first few days after transplantation may be the result of a reduction in the steroid dose during that period. Then, the CL significantly decreased with POD, which was presumably related to reduced bile formation/excretion or hepatic cytochrome P450 metabolism due to transplant- or chemotherapy-related toxicity. Therefore, to avoid cyclosporine-related toxicity, adjustment of the intravenously administered dose is necessary during the early allo-HSCT period. In addition, our study found that POD also has a significant impact on V, which was also proven by Joohan’s research [16]. This tendency may be linked to the rebuilding of the recipients’ hematopoietic system and immune system after transplantation.
At present, the effect of renal function on cyclosporine CL is still controversial. No significant effect was discovered in children after renal and stem cell transplantation [22, 46]. Ni et al. [7] and Fanta et al. [32] reported a significant effect of serum creatinine on CL/F in both renal pretransplant and stem cell transplantation juvenile patients. Similarly, our study showed that the eGFR, which is a more accurate indicator than serum creatinine to reflect renal function, significantly affected cyclosporine clearance in Chinese juvenile allo-HSCT patients. A study in patients with kidney disease reported that renal dysfunction could decrease the activities of hepatic drug-metabolism enzymes, including CYP3A4 [47]. Thus, we speculate that renal dysfunction might decrease the CL of CsA by reducing the activity of CYP3A.
In fact, we also tried stricter statistical standards: P < 0.01 for forward inclusion and P < 0.001 for backward elimination in the modeling process. As a result, only three covariates were retained that had significant impacts on CL, body weight, POD, and coadministration of TAF, while only body weight had significant impacts on V. However, to maximize future research, some possible influencing factors in the new research design could be concerning, so we finally adopted P < 0.05 and P < 0.01 as the criteria for covariate inclusion and elimination, respectively.
Finally, the current study has some limitations. First, only C0, the current common practice for CsA drug monitoring, was used in this population pharmacokinetic analysis. However, the CsA concentration measured at 2 h post dose (C2) has been reported to be better related to the area under the curve (AUC) in transplanted patients, which is considered to be the best marker of drug exposure for predicting the efficacy of drug therapy [48, 49]. Second, CsA concentrations were detected using immunoassays, which may overestimate CsA levels due to cross-reactivity compared with high-performance liquid chromatography–tandem mass spectrometry or high-performance liquid chromatography [50]. Third, not all of the mutation sites studied in the present research were genotyped successfully in each sample because of the quality of DNA or detection method, which may reduce the chance of finding genotype-related covariants. Fourth, the number of samples was not sufficient, and studies with larger sample sizes are needed in the future.
In conclusion, a population pharmacokinetic model of cyclosporine was developed in Chinese children receiving allo-HSCT. Body weight, POD, the CYP3A4*1 G genotype, eGFR and coadministration of TAF significantly influenced the cyclosporine CL, and weight and POD significantly influenced the cyclosporine V. These findings might be useful for physicians in optimizing the dosage of intravenously administered cyclosporine in children.
Acknowledgements
The authors gratefully acknowledge financial support from the National Natural Science Foundation of China (Grant No. 8177131171) and the Beijing Municipal Natural Science Foundation (Grant No. 7192218).
Author contributions
TFL, LH, WYF, and CFW designed the research study. TFL, LH, XML, XXL performed the research study. TFL, XLM and LH participated in patient enrollment and genotypic and data collection. TFL, LH, and XLM analyzed the data. TFL wrote the manuscript, which was critically revised by the other author.
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Wan-yu Feng, Email: renminyaojike@sina.com.
Chun-fu Wu, Email: wucf@syphu.edu.cn.
References
- 1.Holtick U, Chemnitz JM, Hallek M, Scheid C. Allogeneic haematopoietic stem cell transplantation—an overview. Klin Monbl Augenheilkd. 2015;232:641–6. doi: 10.1055/s-0035-1545939. [DOI] [PubMed] [Google Scholar]
- 2.Bouchlaka MN, Redelman D, Murphy WJ. Immunotherapy following hematopoietic stem cell transplantation: potential for synergistic effects. Immunotherapy. 2010;2:399–418. doi: 10.2217/imt.10.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi SW, Reddy P. Current and emerging strategies for the prevention of graft-versus-host disease. Nat Rev Clin Oncol. 2014;11:536–47. doi: 10.1038/nrclinonc.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Citterio F. Evolution of the therapeutic drug monitoring of cyclosporine. Transplant Proc. 2004;36:420s–5s. doi: 10.1016/j.transproceed.2004.01.054. [DOI] [PubMed] [Google Scholar]
- 5.Zheng QS, Li LJ. Pharmacometrics: a quantitative tool of pharmacological research. Acta Pharmacol Sin. 2012;33:1337–8. doi: 10.1038/aps.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarem S, Li J, Barriere O, Litalien C, Theoret Y, Lapeyraque AL, et al. Bayesian approach for the estimation of cyclosporine area under the curve using limited sampling strategies in pediatric hematopoietic stem cell transplantation. Theor Biol Med Model. 2014;11:39. doi: 10.1186/1742-4682-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni SQ, Zhao W, Wang J, Zeng S, Chen SQ, Jacqz-Aigrain E, et al. Population pharmacokinetics of ciclosporin in Chinese children with aplastic anemia: effects of weight, renal function and stanozolol administration. Acta Pharmacol Sin. 2013;34:969–75. doi: 10.1038/aps.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao XR, Xia XY, Zhang J, Tong LY, Zhang W, Zhou X, et al. CYP3A4 *18B and CYP3A5 *3 polymorphisms contribute to pharmacokinetic variability of cyclosporine among healthy Chinese subjects. Eur J Pharm Sci. 2015;76:238–44. doi: 10.1016/j.ejps.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Li JL, Fu Q, Wang XD, Liu LS, Wang CX, et al. Associations of ABCB1, NFKB1, CYP3A, and NR1I2 polymorphisms with cyclosporine trough concentrations in Chinese renal transplant recipients. Acta Pharmacol Sin. 2013;34:555–60. doi: 10.1038/aps.2012.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cvetkovic M, Zivkovic M, Bundalo M, Gojkovic I, Spasojevic-Dimitrijeva B, Stankovic A, et al. Effect of age and allele variants of CYP3A5, CYP3A4, and POR genes on the pharmacokinetics of cyclosporin A in pediatric renal transplant recipients from Serbia. Ther Drug Monit. 2017;39:589–95. doi: 10.1097/FTD.0000000000000442. [DOI] [PubMed] [Google Scholar]
- 11.Lampen A, Christians U, Bader A, Hackbarth I, Sewing KF. Drug interactions and interindividual variability of ciclosporin metabolism in the small intestine. Pharmacology. 1996;52:159–68. doi: 10.1159/000139380. [DOI] [PubMed] [Google Scholar]
- 12.Crettol S, Venetz JP, Fontana M, Aubert JD, Pascual M, Eap CB. CYP3A7, CYP3A5, CYP3A4, and ABCB1 genetic polymorphisms, cyclosporine concentration, and dose requirement in transplant recipients. Ther Drug Monit. 2008;30:689–99. doi: 10.1097/FTD.0b013e31818a2a60. [DOI] [PubMed] [Google Scholar]
- 13.Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu Rev Pharmacol Toxicol. 1999;39:361–98. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 14.Sakaeda T, Nakamura T, Okumura K. Pharmacogenetics of MDR1 and its impact on the pharmacokinetics and pharmacodynamics of drugs. Pharmacogenomics. 2003;4:397–410. doi: 10.1517/phgs.4.4.397.22747. [DOI] [PubMed] [Google Scholar]
- 15.Qiu XY, Jiao Z, Zhang M, Zhong LJ, Liang HQ, Ma CL, et al. Association of MDR1, CYP3A4*18B, and CYP3A5*3 polymorphisms with cyclosporine pharmacokinetics in Chinese renal transplant recipients. Eur J Clin Pharmacol. 2008;64:1069–84. doi: 10.1007/s00228-008-0520-8. [DOI] [PubMed] [Google Scholar]
- 16.Song J, Kim MG, Choi B, Han NY, Yun HY, Yoon JH, et al. CYP3A5 polymorphism effect on cyclosporine pharmacokinetics in living donor renal transplant recipients: analysis by population pharmacokinetics. Ann Pharmacother. 2012;46:1141–51. doi: 10.1345/aph.1R004. [DOI] [PubMed] [Google Scholar]
- 17.Dong ZL, Li H, Chen QX, Hu Y, Wu SJ, Tang LY, et al. Effect of CYP3A4*1G on the fentanyl consumption for intravenous patient-controlled analgesia after total abdominal hysterectomy in Chinese Han population. J Clin Pharm Ther. 2012;37:153–6. doi: 10.1111/j.1365-2710.2011.01268.x. [DOI] [PubMed] [Google Scholar]
- 18.Hu L, Zhuo W, He YJ, Zhou HH, Fan L. Pharmacogenetics of P450 oxidoreductase: implications in drug metabolism and therapy. Pharmacogenet Genomics. 2012;22:812–9. doi: 10.1097/FPC.0b013e328358d92b. [DOI] [PubMed] [Google Scholar]
- 19.Lunde I, Bremer S, Midtvedt K, Mohebi B, Dahl M, Bergan S, et al. The influence of CYP3A, PPARA, and POR genetic variants on the pharmacokinetics of tacrolimus and cyclosporine in renal transplant recipients. Eur J Clin Pharmacol. 2014;70:685–93. doi: 10.1007/s00228-014-1656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniele C, Katja B, Slager SL, Skibola CF, Silvia DS, Yolanda B, et al. A comprehensive study of polymorphisms in the ABCB1, ABCC2, ABCG2, NR1I2 genes and lymphoma risk. Int J Cancer J Int Du Cancer. 2012;131:803–12. doi: 10.1002/ijc.26436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, LeCluyse EL. Role of orphan nuclear receptors in the regulation of drug-metabolising enzymes. Clin Pharmacokinet. 2003;42:1331–57. doi: 10.2165/00003088-200342150-00003. [DOI] [PubMed] [Google Scholar]
- 22.Kim MG, Kim IW, Choi B, Han N, Yun HY, Park S, et al. Population pharmacokinetics of cyclosporine in hematopoietic stem cell transplant patients: consideration of genetic polymorphisms. Ann Pharmacother. 2015;49:622–30. doi: 10.1177/1060028015577798. [DOI] [PubMed] [Google Scholar]
- 23.Grinyo J, Vanrenterghem Y, Nashan B, Vincenti F, Ekberg H, Lindpaintner K, et al. Association of four DNA polymorphisms with acute rejection after kidney transplantation. Transpl Int. 2008;21:879–91. doi: 10.1111/j.1432-2277.2008.00679.x. [DOI] [PubMed] [Google Scholar]
- 24.Lupianez CB, Villaescusa MT, Carvalho A, Springer J, Lackner M, Sanchez-Maldonado JM, et al. Common genetic polymorphisms within NFkappaB-related genes and the risk of developing invasive Aspergillosis. Front Microbiol. 2016;7:1243. doi: 10.3389/fmicb.2016.01243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shu WY, Li JL, Wang XD, Huang M. Pharmacogenomics and personalized medicine: a review focused on their application in the Chinese population. Acta Pharmacol Sin. 2015;36:535–43. doi: 10.1038/aps.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Z, Xu Q, Qiu X, Jiao Z, Zhang M, Zhong M. FOXP3rs3761548 polymorphism is associated with tacrolimus-induced acute nephrotoxicity in renal transplant patients. Eur J Clin Pharmacol. 2017;73:39–47. doi: 10.1007/s00228-016-2140-z. [DOI] [PubMed] [Google Scholar]
- 27.Shu W, Guan S, Yang X, Liang L, Li J, Chen Z, et al. Genetic markers in CYP2C19 and CYP2B6 for prediction of cyclophosphamide’s 4-hydroxylation, efficacy and side effects in Chinese patients with systemic lupus erythematosus. Br J Clin Pharmacol. 2016;81:327–40. doi: 10.1111/bcp.12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao W, Elie V, Roussey G, Brochard K, Niaudet P, Leroy V, et al. Population pharmacokinetics and pharmacogenetics of tacrolimus in de novo pediatric kidney transplant recipients. Clin Pharmacol Ther. 2009;86:609–18. doi: 10.1038/clpt.2009.210. [DOI] [PubMed] [Google Scholar]
- 29.Hope WW, Smith PB, Arrieta A, Buell DN, Roy M, Kaibara A, et al. Population pharmacokinetics of micafungin in neonates and young infants. Antimicrob Agents Chemother. 2010;54:2633–7. doi: 10.1128/AAC.01679-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson BJ, Holford NH. Mechanism-based concepts of size and maturity in pharmacokinetics. Annu Rev Pharmacol Toxicol. 2008;48:303–32. doi: 10.1146/annurev.pharmtox.48.113006.094708. [DOI] [PubMed] [Google Scholar]
- 31.Brendel K, Comets E, Laffont C, Laveille C, Mentre F. Metrics for external model evaluation with an application to the population pharmacokinetics of gliclazide. Pharm Res. 2006;23:2036–49. doi: 10.1007/s11095-006-9067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fanta S, Jonsson S, Backman JT, Karlsson MO, Hoppu K. Developmental pharmacokinetics of ciclosporine population pharmacokinetic study in paediatric renal transplant candidates. Br J Clin Pharmacol. 2007;64:772–84. doi: 10.1111/j.1365-2125.2007.03003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irtan S, Saint-Marcoux F, Rousseau A, Zhang D, Leroy V, Marquet P, et al. Population pharmacokinetics and bayesian estimator of cyclosporine in pediatric renal transplant patients. Ther Drug Monit. 2007;29:96–102. doi: 10.1097/FTD.0b013e3180310f9d. [DOI] [PubMed] [Google Scholar]
- 34.Xue L, Zhang WW, Ding XL, Zhang JJ, Bao JA, Miao LY. Population pharmacokinetics and individualized dosage prediction of cyclosporine in allogeneic hematopoietic stem cell transplant patients. Am J Med Sci. 2014;348:448–54. doi: 10.1097/MAJ.0000000000000337. [DOI] [PubMed] [Google Scholar]
- 35.Dai Y, Iwanaga K, Lin YS, Hebert MF, Davis CL, Huang W, et al. In vitro metabolism of cyclosporine A by human kidney CYP3A5. Biochem Pharmacol. 2004;68:1889–902. doi: 10.1016/j.bcp.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 36.Chen B, Zhang W, Gu Z, Li J, Zhang Y, Cai W. Population pharmacokinetic study of cyclosporine in Chinese renal transplant recipients. Eur J Clin Pharmacol. 2011;67:601–12. doi: 10.1007/s00228-010-0959-2. [DOI] [PubMed] [Google Scholar]
- 37.Fruit D, Rousseau A, Amrein C, Rolle F, Kamar N, Sebbag L, et al. Ciclosporin population pharmacokinetics and Bayesian estimation in thoracic transplant recipients. Clin Pharmacokinet. 2013;52:277–88. doi: 10.1007/s40262-013-0037-x. [DOI] [PubMed] [Google Scholar]
- 38.Rosenbaum SE, Baheti G, Trull AK, Akhlaghi F. Population pharmacokinetics of cyclosporine in cardiopulmonary transplant recipients. Ther Drug Monit. 2005;27:116–22. doi: 10.1097/01.ftd.0000148448.51225.2c. [DOI] [PubMed] [Google Scholar]
- 39.Zhou H, Gao Y, Cheng XL, Li ZD. Population pharmacokinetics of cyclosporine A based on NONMEM in Chinese allogeneic hematopoietic stem cell transplantation recipients. Eur J Drug Metab Pharmacokinet. 2012;37:271–8. doi: 10.1007/s13318-012-0087-8. [DOI] [PubMed] [Google Scholar]
- 40.Lempers VJ, Martial LC, Schreuder MF, Blijlevens NM, Burger DM, Aarnoutse RE, et al. Drug-interactions of azole antifungals with selected immunosuppressants in transplant patients: strategies for optimal management in clinical practice. Curr Opin Pharmacol. 2015;24:38–44. doi: 10.1016/j.coph.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Nara M, Takahashi N, Miura M, Niioka T, Kagaya H, Fujishima N, et al. Effect of itraconazole on the concentrations of tacrolimus and cyclosporine in the blood of patients receiving allogeneic hematopoietic stem cell transplants. Eur J Clin Pharmacol. 2013;69:1321–9. doi: 10.1007/s00228-013-1471-2. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Y, Sheng XY, Xu JY, Bi SS, Lu W, Cui YM. Population pharmacokinetic study of cyclosporine in the hematopoietic stem cell transplant recipients. Int J Clin Pharmacol Ther. 2013;51:568–75. doi: 10.5414/CP201815. [DOI] [PubMed] [Google Scholar]
- 43.Okada A, Ushigome H, Kanamori M, Morikochi A, Kasai H, Kosaka T, et al. Population pharmacokinetics of cyclosporine A in Japanese renal transplant patients: comprehensive analysis in a single center. Eur J Clin Pharmacol. 2017;73:1111–9. doi: 10.1007/s00228-017-2279-2. [DOI] [PubMed] [Google Scholar]
- 44.Wu KH, Cui YM, Guo JF, Zhou Y, Zhai SD, Cui FD, et al. Population pharmacokinetics of cyclosporine in clinical renal transplant patients. Drug Metab Dispos. 2005;33:1268–75. doi: 10.1124/dmd.105.004358. [DOI] [PubMed] [Google Scholar]
- 45.Jacobson PA, Ng J, Green KG, Rogosheske J, Brundage R. Posttransplant day significantly influences pharmacokinetics of cyclosporine after hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2003;9:304–11. doi: 10.1016/S1083-8791(03)00076-4. [DOI] [PubMed] [Google Scholar]
- 46.Willemze AJ, Cremers SC, Schoemaker RC, Lankester AC, den Hartigh J, Burggraaf J, et al. Ciclosporin kinetics in children after stem cell transplantation. Br J Clin Pharmacol. 2008;66:539–45. doi: 10.1111/j.1365-2125.2008.03217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nolin TD, Naud J, Leblond FA, Pichette V. Emerging evidence of the impact of kidney disease on drug metabolism and transport. Clin Pharmacol Ther. 2008;83:898–903. doi: 10.1038/clpt.2008.59. [DOI] [PubMed] [Google Scholar]
- 48.Ferraresso M, Ghio L, Tirelli S, Pedotti P, Taioli E, Edefonti A, et al. Cyclosporine monitoring in stable, long-term, pediatric kidney transplant recipients: the value of C2 determination. Transplant Proc. 2004;36:685–6. doi: 10.1016/j.transproceed.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Zhao W, Fakhoury M, Jacqz-Aigrain E. Developmental pharmacogenetics of immunosuppressants in pediatric organ transplantation. Ther Drug Monit. 2010;32:688–99. doi: 10.1097/FTD.0b013e3181f6502d. [DOI] [PubMed] [Google Scholar]
- 50.Salm P, Taylor PJ, Rooney F. A high-performance liquid chromatography-mass spectrometry method using a novel atmospheric pressure chemical ionization approach for the rapid simultaneous measurement of tacrolimus and cyclosporin in whole blood. Ther Drug Monit. 2008;30:292–300. doi: 10.1097/FTD.0b013e3181771feb. [DOI] [PubMed] [Google Scholar]