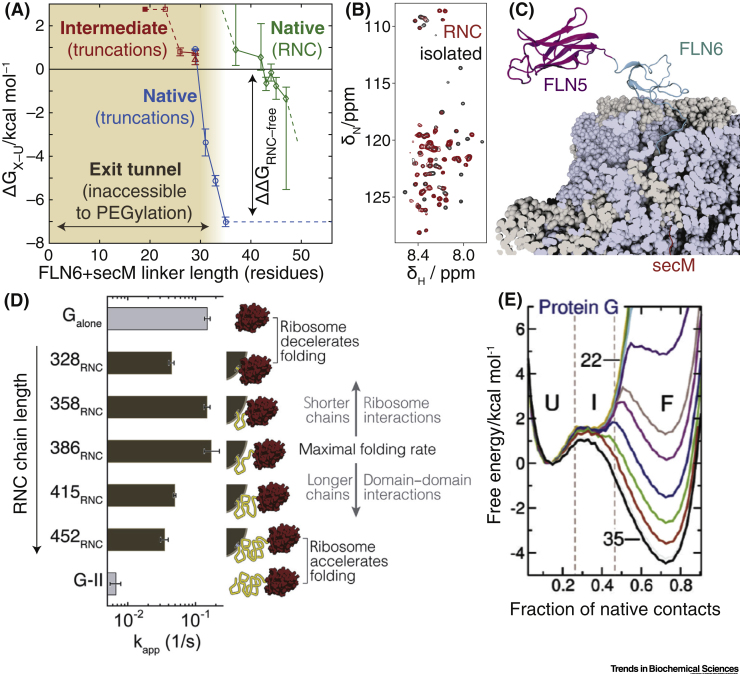
Figure I.
Experimental and Computational Characterisation of Co-translational Folding.
(A) NMR and biochemical characterisation of length-dependent folding in an FLN5+6 ribosome–nascent chain complex (RNC) system, showing the extent of the exit tunnel (shaded yellow) measured by the susceptibility of a cysteine residue in an unfolded RNC to PEGylation (resulting from a covalent reaction with a 5 kDa polyethylene glycol maleimide), and free energies for folding of the tethered FLN5 domain (green) determined via an analysis of NMR resonance intensities [37]. Free energies for folding are also shown for isolated native and intermediate states (associated with the isomerisation of a native-state cis proline) determined using a C-terminal truncation approach and indicate a substantial destabilisation of the native state on the ribosome 14, 37. (B) The 1H,15N NMR correlation spectrum of an FLN5+31 RNC, overlaid with that of an isolated unfolded reference, reveals residue-specific broadenings associated with interactions between the ribosome surface and the disordered nascent polypeptide chain (NC) [37]. (C) Snapshot from an ensemble structure of the FLN5+110 RNC system (comprising the FLN5 domain attached to a 110 amino acid linker corresponding to the subsequent FLN6 domain and the secM arrest peptide), determined using molecular dynamics simulations restrained using experimental values of NMR chemical shifts [37]. (D) Apparent folding rate of the G domain of EF-G, measured for isolated G and G-II domains and for varying lengths of G-II RNCs, through repeated force-ramp cycles using optical tweezers [88]. (E) Computational modelling of length-dependent free energy landscapes using coarse-grained molecular dynamics simulations, showing in this case the progressive stabilisation of the folded state ‘F’ of protein G from linker lengths of 22–35 residues [101]. Reprinted (adapted), with permission, from [101]. Abbreviation: PPM. Parts per million.