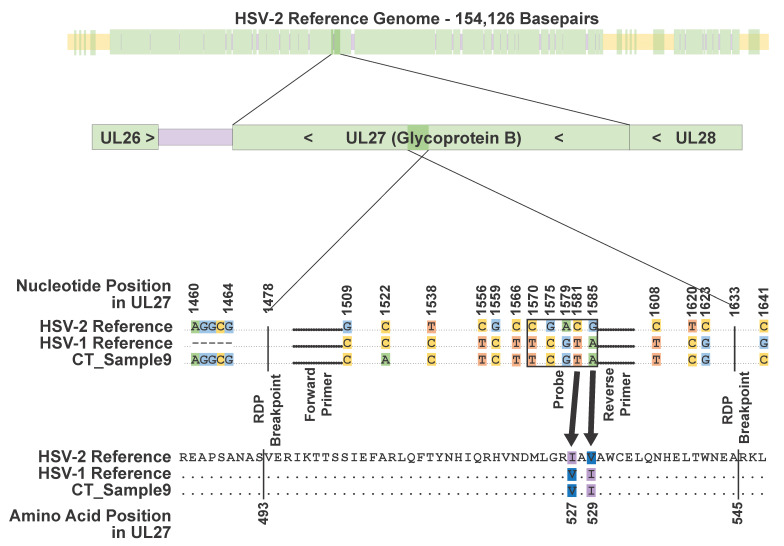
Figure 1.
Nucleotides involved in the recombination event in UL27. The top bar shows the location of the UL27 gene (shown in dark colors) within the HSV-2 genome (faded out). Genic regions in the HSV-2 genome are shown in green, while intragenic sequences outside the terminal and internal repeat regions are in purple. Intragenic regions within the terminal and internal repeat regions are in yellow. The second bar shows UL27 in its genic neighborhood with the recombinant region highlighted. UL27 is in reverse orientation in the HSV genome. In the alignment, nucleotides that are identical in all sequences are represented by dots, while nucleotides that differ among sequences are represented by letters. Dots shown in bold represent the locations of the forward and reverse primers, as labeled. The probe region is boxed. Recombination event breakpoints, as identified by the recombination detection program (RDP), are labeled. Non-synonymous differences between HSV-1 and HSV-2 are indicated with arrows pointing to the corresponding amino acid changes.