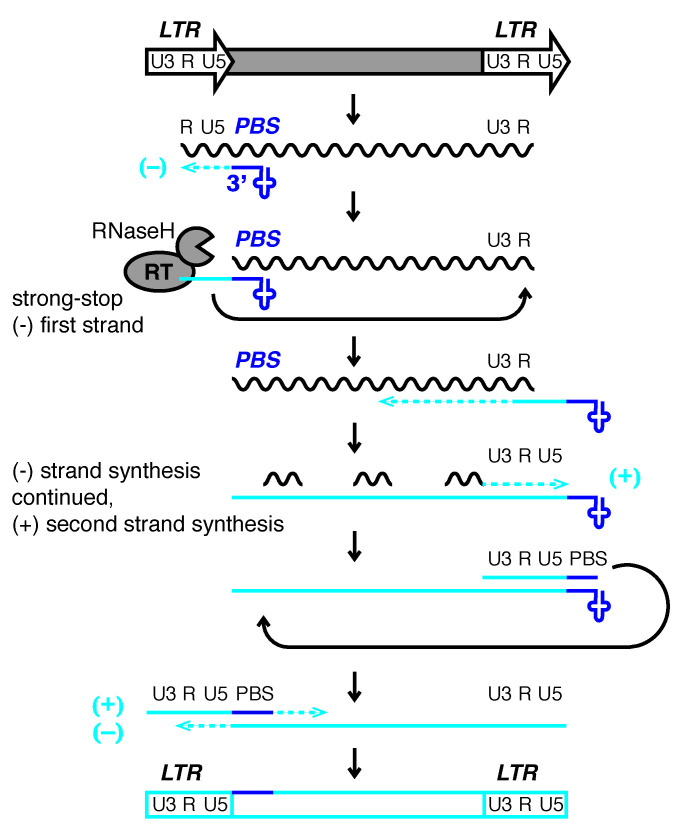
Figure 1.
Model of reverse transcription of long terminal repeat (LTR)-retrotransposons and -viruses. LTRs encode promoter elements and termination signals. The RNA transcript contains a region repeated at either end (R), a 5′ unique segment (U5), and a segment only included at the 3′-end of the RNA (U3). The 3′-end of cellular tRNAs (blue cloverleaf) primes reverse transcription by hybridizing to the primer binding site (PBS). While this segment is being copied into first-strand cDNA (light blue line), also called minus (−) strong stop DNA, the RNaseH activity of reverse transcriptase (RT) degrades the template RNA. The elongating cDNA is transferred to the 3′-end of the retrotransposon transcript hybridizing to the R region. The remaining RNA is partially degraded by RNaseH leaving behind primers for second-strand, plus (+) cDNA synthesis. In Retroviridae, the plus strand PBS is a copy of the tRNA primer, while the minus strand is a copy of the original PBS sequence. After another transfer event, first (−) and second (+) strand synthesis are completed to result in a full-length, double-stranded retroviral DNA that will be integrated into the host genome.