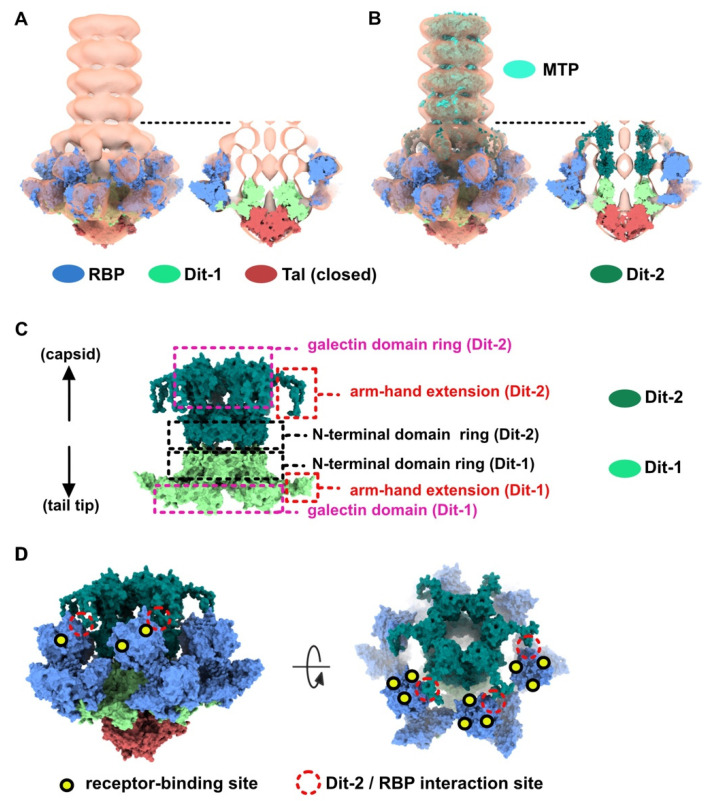
Figure 2.
Topological model of the p2 baseplate resting state in virions. (A) 3D reconstruction of p2 virion’s tail tip and baseplate (EMD-1699) and surface representation of p2 receptor-binding protein (RBPs), Dit-1 and Tal crystal structures (PDB ID 2WZP) fitted in the map. The RBPs point towards the capsid and the Tal trimer is closed. An overall view (left) and a central section (right) are shown. The dotted line indicates the junction between the tail (MTPs) and the baseplate. (B) Same views as in A with the model of the second Dit hexamer (Dit-2) fitted in the map (surface representation). Four hexameric rings of the staphylococcal phage 80α major tail proteins (MTPs) are also shown in the tail as model structures for p2 MTPs (PDB ID 6V8I) [20]. (C) Topological model of the Dit-1 and Dit-2 assembly highlighting the organization of their N-terminal domain, arm-hand extension and galectin domain. (D) Topological model of the baseplate resting state (RBP, Tal, Dit-1, Dit-2) in p2 virions (PDB ID 6ZJJ). Side and tilted views are shown. The red dotted circles indicate the interaction between the Dit-2 arm-hand extension and one RBP. The receptor-binding sites in trimeric RBPs are indicated with yellow/black dots.