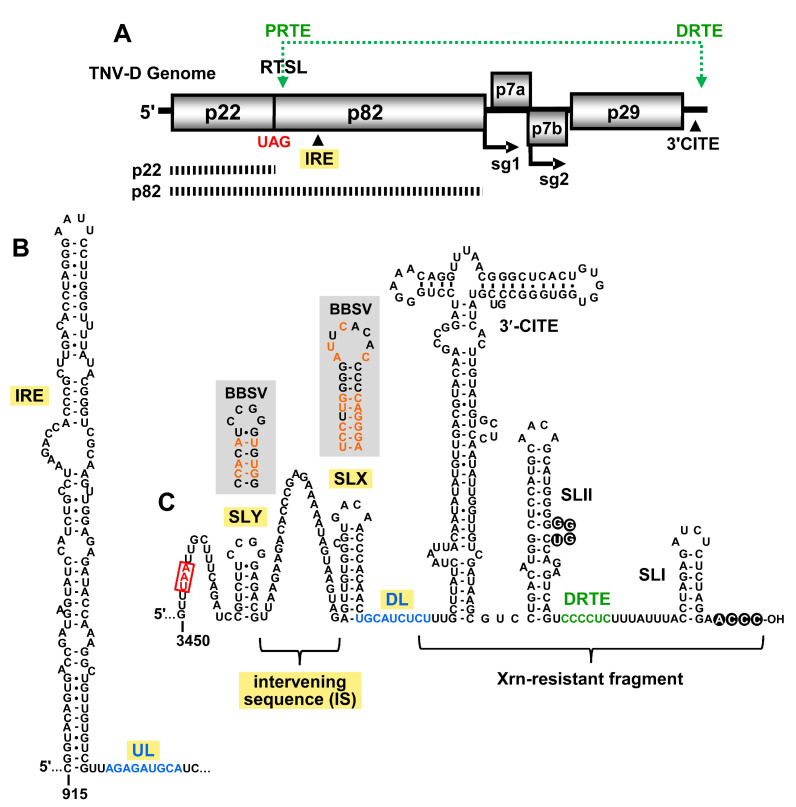
Figure 1.
Tobacco necrosis virus strain D (TNV-D) genome and the predicted RNA secondary structures. (A) A cartoon representation of the TNV-D genome with the encoded proteins represented by grey boxes. The UAG stop codon for p22 is indicated in red. Subgenomic mRNA1 and 2 initiation sites (sg1 and sg2) are shown below the genome. The location of the 3′ cap independent translational enhancer (3′CITE) and the internal replication element (IRE) are also shown. The black dashed bars below the genome correspond to the p22 protein and the p82 readthrough product. The green double-headed arrow shows a long-range RNA–RNA interaction between the proximal readthrough element (PRTE; located within the readthrough stem–loop, RTSL) and the distal readthrough element (DRTE; in the 3′UTR) that is necessary for efficient readthrough. (B) Mfold-predicted RNA secondary structure of the TNV-D internal replication element (IRE), located within the coding region of p82. The 3′-adjacent upstream linker (UL) sequence is shown in blue. (C) Predicted RNA secondary structure of the 3′UTR in the TNV-D genome based on comparative structural analysis, compensatory mutational analysis, and structure probing data [2,5,6,7]. The UAA stop codon of the p29 capsid protein is boxed and highlighted in red, and the portion of the 3′UTR that accumulates as an Xrn-resistant fragment in infections is delineated. The 3′CITE and relevant stem–loop (SL) structures are labelled, as are the intervening sequence (IS), downstream linker (DL; blue) and DRTE (green). The grey boxes show the corresponding secondary structures of SLX and SLY for Beet black scorch virus (BBSV), with orange nucleotides representing the sequence changes when compared with TNV-D.