Figure 1.

Methods overview. A. Electrode locations. Each dot reflects the location of a single electrode implanted in the brain of a Dataset 1 patient. A held-out recording location from one patient is indicated in red, and the patient’s remaining electrodes are indicated in black. The electrodes from the remaining patients are colored by k-means cluster (computed using the full-brain correlation model shown in Panel D). B. Radial basis function kernel. Each electrode contributed by the patient (black) weights on the full set of locations under consideration (all dots in Panel A, defined as 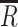 in the text). The weights fall off with positional distance (in MNI152 space) according to an RBF. C. Per-patient correlation matrices. After computing the pairwise correlations between the recordings from each patient’s electrodes, we use RBF-weighted averages to estimate correlations between all locations in
in the text). The weights fall off with positional distance (in MNI152 space) according to an RBF. C. Per-patient correlation matrices. After computing the pairwise correlations between the recordings from each patient’s electrodes, we use RBF-weighted averages to estimate correlations between all locations in  . We obtain an estimated full-brain correlation matrix using each patient’s data. D. Merged correlation model. We combine the per-patient correlation matrices (Panel C) to obtain a single full-brain correlation model that captures information contributed by every patient. Here, we have sorted the rows and columns to reflect k-means clustering labels (using k = 7; Yeo et al. 2011), whereby we grouped locations based on their correlations with the rest of the brain (i.e., rows of the matrix displayed in the panel). The boundaries denote the cluster groups. The rows and columns of Panel C have been sorted using the Panel D-derived cluster labels. E. Reconstructing activity throughout the brain. Given the observed recordings from the given patient (shown in black; held-out recording is shown in blue), along with a full-brain correlation model (Panel D), we use equation (12) to reconstruct the most probable activity at the held-out location (red).
. We obtain an estimated full-brain correlation matrix using each patient’s data. D. Merged correlation model. We combine the per-patient correlation matrices (Panel C) to obtain a single full-brain correlation model that captures information contributed by every patient. Here, we have sorted the rows and columns to reflect k-means clustering labels (using k = 7; Yeo et al. 2011), whereby we grouped locations based on their correlations with the rest of the brain (i.e., rows of the matrix displayed in the panel). The boundaries denote the cluster groups. The rows and columns of Panel C have been sorted using the Panel D-derived cluster labels. E. Reconstructing activity throughout the brain. Given the observed recordings from the given patient (shown in black; held-out recording is shown in blue), along with a full-brain correlation model (Panel D), we use equation (12) to reconstruct the most probable activity at the held-out location (red).
