Fig. 6.
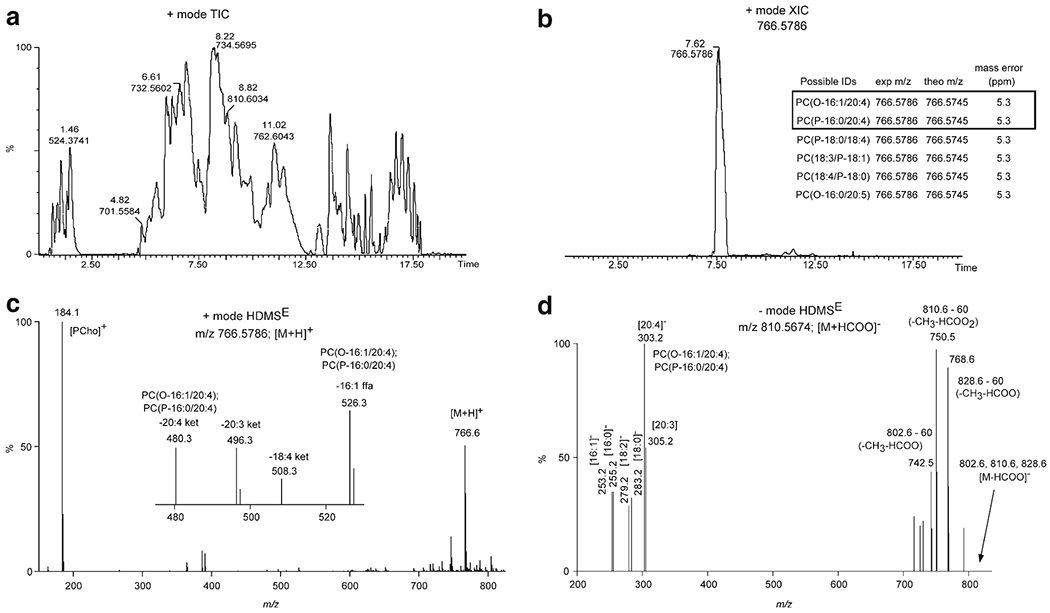
UPLC-HDMSE structure confirmation for PCe36:5. (a) Positive ion mode total ion chromatogram (TIC) of mouse lung total lipid extract. Retention time (min) and m/z value denoted for several of the most abundant chromatographic peaks. (b) Extracted ion chromatogram (XIC) for the m/z value at 766.5786. The inset table lists the possible lipid structures for accurate mass (<6.5 ppm) database searching from LIPID MAPS. PC(O-16:1/22:4) and PC(P-16:0/22:4) were the structures confirmed based on UPLC-HDMSE. (c) Positive ion mode HDMSE high-collision energy scan for m/z 766.5786 ([M + H]+ ion for PCe36:5). The inset mass spectrum is the zoomed in region for m/z 470–530. The m/z value and associated product ion or neutral loss is listed next to the peak. (d) Negative ion mode HDMSE high-collision energy scan for m/z 810.5674 ([M + HCOO]− ion for PCe36:5). The m/z value and associated product ion or neutral loss is listed next to the peak.
