Fig. 3.
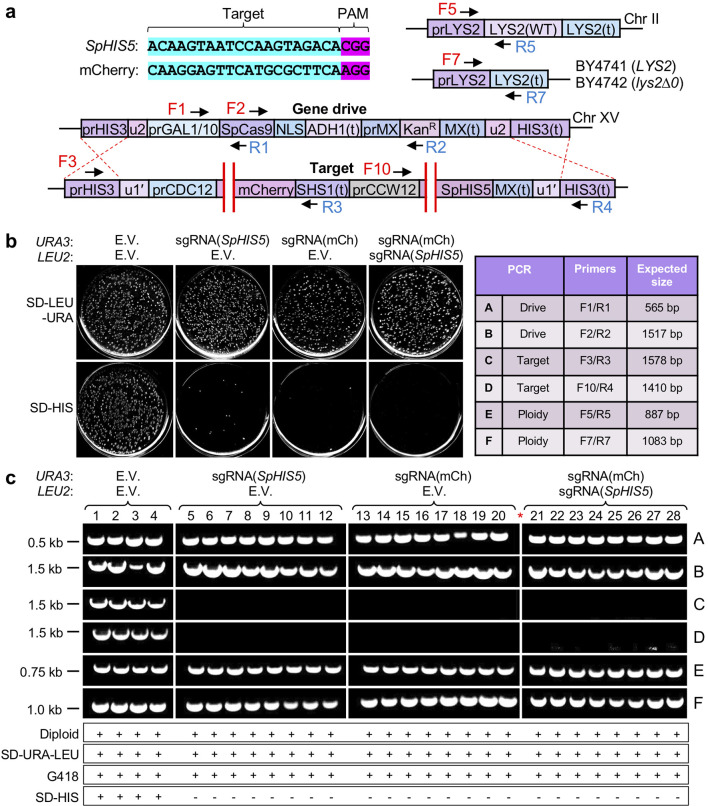
Use of alternative sgRNAs within the target allele allows for successful CRISPR GD action. (a) Schematic of a modified drive/target system that included target sites within the mCherry and SpHIS5 coding sequences. Cross-over between the MX(t) DNA is also possible upstream of the (u1’) and (u2) sequences, but is not illustrated for clarity. (b) Two high-copy plasmids (URA3-based, pGF-V2153, pGF-V2159; LEU2-based, pGF-V2152) were included within the drive strain GFY-2383. EV, empty pRS425 or pRS426 vectors. Following activation in galactose, diploids were plated as in Fig. 1 using SD-LEU-URA and SD-HIS medium. Individual plate images (full plates) were edited for contrast. (c) Diagnostic PCRs of clonal diploid isolates from (c). Top right: table of primers chosen for analysis and the expected product sizes. Horizontal white lines designate separate DNA gels. Vertical white lines are included for clarity; red asterisk, a separate DNA gel was used for samples 21–28.