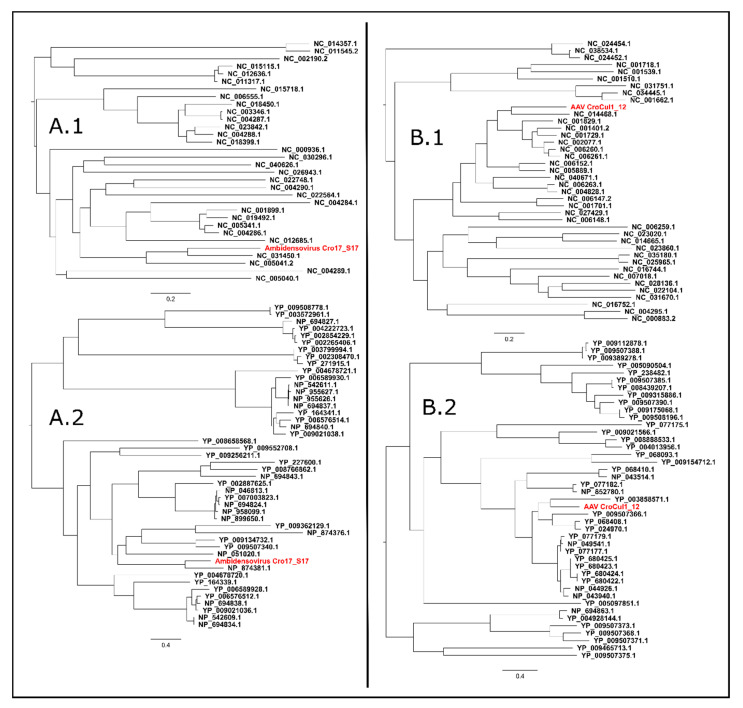
Figure 2.
Phylogenetic clustering of Ambidensovirus Cro17_S17 (A) and Adeno-associated virus AAV CroCul1_12 (B) based on the complete genome nucleotide (A.1,B.1) and the NS1/Rep amino acid contexts. All phylogenetic trees were inferred using IQ-TREE, best-fitting phylogenetic models were selected automatically based on the Bayesian information criterion; all trees were visualized using FigTree and rooted at mid-point. The complete genome phylogenetic trees were constructed based on the complete genome sequences matching Densovirinae (A.1) and Parvovirinae (B.1) taxons in NCBI RefSeq using phylogenetic models GTR+F+R4 and GTR+F+R6, respectively. The NS1/Rep protein phylogenetic trees were constructed based on NS1 and Rep proteins sequences from the NCBI Non-redundant protein dataset matching Densovirinae (A.2) and Parvovirinae (B.2) taxons, using phylogenetic models VT+F+R4 and VT+F+R9.