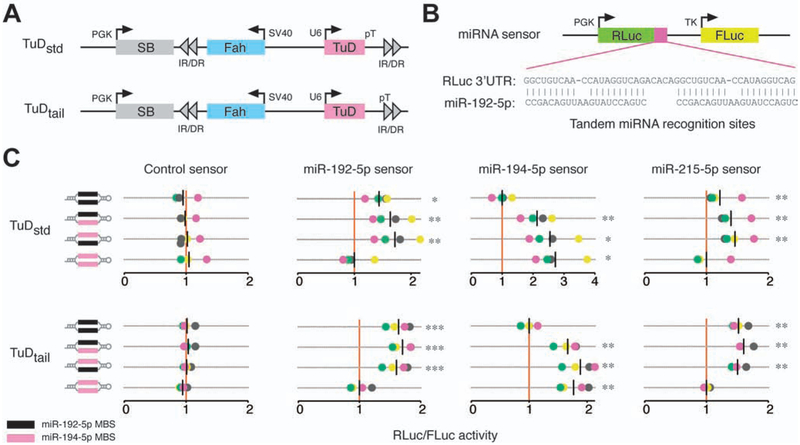
Figure 1.
Inhibition of microRNA activity via tough decoy expression. (A) Fah plasmids expressing TuDs from a U6 promoter as standard hairpins (TuDstd) or with extended 3’ sequences (TuDtail). Plasmids encode sleeping beauty (SB) transposase and inverted repeat/direct repeat (IR/DR) sequences for genomic integration. (B) MiRNA sensor plasmids encoding a Renilla luciferase (RLuc) cDNA with miRNA binding sites in the 3’UTR and a non-targeted firefly luciferase (FLuc) cDNA. (C) Dual luciferase assays in mouse Hepa 1–6 cells co-transfected with Fah plasmids encoding TuDs directed against the miRNA-194/192/215 family and miRNA sensor plasmids. The RLuc/FLuc ratios for miRNA sensors are scaled such that the mean of the sensor-specific control TuD is one (red vertical line). Assays were performed in quadruplicate; data points are colored by replicate. Black vertical lines indicate the mean luciferase ratio of each TuD. *P<0.05; **P<0.01; ***P<0.001, repeated measures ANOVA with Dunnet’s test for multiple comparisons.