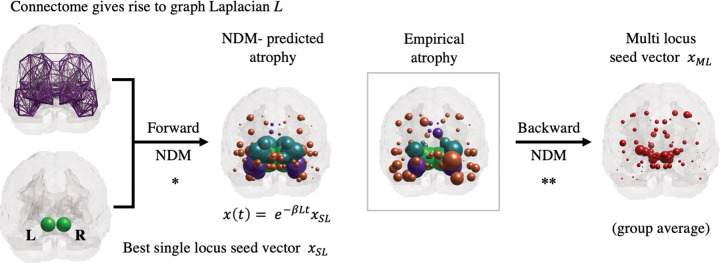
Figure 1.
Overview of forward and backward NDM. NDM is an established graph-theoretical model for neuropathology spread. Each node of the graph is associated with a grey matter region specified by a brain atlas. The connectome gives rise to the graph-Laplacian L. To analytically predict the value of the regional atrophy vector at a given point , one must specify the initial state of the system. It is customary in the forward NDM to test all possible single-locus seed vectors to find the one that best predicts the empirical atrophy (*). The backward NDM is a recently developed algorithm that can infer more predictive and sophisticated initial states , with multiple loci and distinct entry values (**). Brain renderings show the coronal view, with pathology loads proportional to the sphere diameters (values scaled for improved visualization). When applied to individual patients, the backward NDM gives rise to incipient patterns that are more heterogeneous than generally appreciated.