Abstract
The Envelope (E) protein of SARS-CoV-2 is the most enigmatic protein among the four structural ones. Most of its current knowledge is based on the direct comparison to the SARS E protein, initially mistakenly undervalued and subsequently proved to be a key factor in the ER-Golgi localization and in tight junction disruption.
We compared the genomic sequences of E protein of SARS-CoV-2, SARS-CoV and the closely related genomes of bats and pangolins obtained from the GISAID and GenBank databases. When compared to the known SARS E protein, we observed a significant difference in amino acid sequence in the C-terminal end of SARS-CoV-2 E protein.
Subsequently, in silico modelling analyses of E proteins conformation and docking provide evidences of a strengthened binding of SARS-CoV-2 E protein with the tight junction-associated PALS1 protein. Based on our computational evidences and on data related to SARS-CoV, we believe that SARS-CoV-2 E protein interferes more stably with PALS1 leading to an enhanced epithelial barrier disruption, amplifying the inflammatory processes, and promoting tissue remodelling. These findings raise a warning on the underestimated role of the E protein in the pathogenic mechanism and open the route to detailed experimental investigations.
Keywords: SARS-CoV-2, COVID-19, Envelope protein, Tight junctions, PALS1
Severe acute respiratory syndrome 2, known as Coronavirus disease 2019 (Covid-19) is caused by a Betacoronavirus named SARS-CoV-2 virus. To date, Covid-19 associated pneumonia accounts (as of 14th July 2020, source: www.who.int, “COVID-19 Situation report - 176”) almost 13 Million confirmed cases and more than 550′000 deaths since its first reported discovery in the Hubei province in China in December 2019 [1].
Bat and Malayan pangolin coronavirus genomes show a high identity percentage to SARS-CoV-2, suggesting these animal species as possible reservoir hosts for SARS-CoV-2 related viruses before adaptation to humans [2]. This hypothesis is well supported by recent findings in genome analysis. In this sense, Spike (S) protein investigation has highlighted a common pattern with the bat and pangolin orthologues, as well as a peculiar motif likely acquired during human adaptation of the SARS-CoV-2 [3].
Coronavirus infection starts with inhalation of droplets containing virus that invades the epithelial cells by using angiotensin converting enzyme 2 (ACE2) [4], or other cell components like integrins, as targets of the SARS-CoV2 S protein [5,6]. Viral replication in Type II alveolar epithelial cells leads to severe modifications of the innate immune response [7]. Lungs are rapidly compromised following direct damage of the pulmonary tissue mainly through dysregulation of the immune mediators that enhance the influx of monocytes and neutrophils in the infected tissue [4]. Furthermore, pro-inflammatory cytokine storm affects virus replication and increases its diffusion to nearby cells.
SARS-CoV-2 virus shows a single strand, positive-sense RNA genome of slightly less than 30 kb in length where at 3’ end are located the genes coding for the four structural proteins: spike (S), membrane (M), envelope (E), and nucleocapsid (N) [1]. As an enveloped virus, S, M and E proteins encounter cellular membrane at the initiation of infection and during the replication cycle and are involved in the budding of the mature virions [8]. Whereas Spike protein has been immediately investigated [3,[9], [10], [11], [12]], less information has been collected on the other surface proteins. Indeed, as occurred with the previously studied Betacoronavirus, the role of E protein and its involvement with the host adverse effects was wrongly underestimated. In contrast to S, E protein of the SARS-CoV-2 has not been thoroughly studied yet. During the initial outbreak, efforts were put into the identification of epitopes with potential cross-protective role [13] while most of the information about its structure and function derive from SARS-CoV experimental studies. While E protein is not represented in the mature virions up to levels of S or M proteins [8,14], it is abundantly expressed inside the infected cell and actively involved in the pathogenic viral mechanisms [15].
SARS-CoV E protein is the smallest among structural proteins (76 amino acids), organized in three main domains: a short (approx. 8–10 amino acids) luminal oriented N-terminal domain, a long α-helical transmembrane domain composed of ≈22 amino acid residues and a cytoplasmically oriented C-terminal domain [16,17].
Homologous assembling of the E protein contributes to create a pentameric channel with its transmembrane domain that directly alters virus replication [18].
Conversely, monomeric E protein affects the host’s intracellular activities through C-terminal end domain, which is predicted to have a β-coil-β structure, leading to its localization in the endoplasmic reticulum, Golgi and the ER-Golgi intermediate compartment [[17], [18], [19]]. Additional targeting information at the N-terminal domain of the E protein would ensure the maintaining of the Golgi complex targeting [19]. Interestingly, last four hydrophobic aminoacidic residues of the C-terminal domain (DLLV) have been indicated to be a PDZ (PSD-95, Dlg, ZO-1 homology) binding motif, as previously reported by Teoh et al. [20]. Particularly, DLLV motif has been demonstrated to compete with Crumbs cell polarity complex component 1 (CRB1) for binding to PALS1 PDZ domain [20]. PALS1 is a cellular protein involved in maintaining tight junctions between epithelial cells also via the interaction with PALS1-associated tight junction (PATJ) protein. The Crumbs-PALS1-PATJ complex is fundamental for the development and maintenance of apical-basal polarity of epithelial cells [21]. Therefore, interactions between the SARS E protein and PALS1 induced relocation of PALS1 to the virus assembly site and disrupted tight junctions promoting virus spread. Little information has been collected yet on SARS-CoV-2 E protein and mainly focused on the sequences conserved from SARS, suggesting its potential interaction with bromodomain proteins [22]. Conversely, aim of this work is to identify the potential implications of sequence dissimilarities between the previous SARS-CoV and SARS-CoV-2.
2. Materials and methods
2.1. Data collection and multiple sequence alignment
Reference coronavirus genomes were obtained either from NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) or from GISAID (https://www.gisaid.org/) databases as suggested from the recent discoveries on the proximal origins of SARS-CoV-2 [2,3] (Detailed accession numbers in Supplementary Table 1). Pangolin genomes were pooled in two groups, as shown by Lam et al. phylogenetic analyses [2] and only the consensus genome for each one of the two groups was considered. The Envelope proteins (E) from all genomes were extracted and multiple sequence alignment performed with the MAFFT algorithm (v7.450) in Geneious Prime (version 11.0.4) [23,24]. Pairwise sequence identities were also calculated using Geneious Prime.
2.2. 3D homology modeling of SARS-CoV-2 E
The amino acid sequence of the SARS-CoV-2 Envelope protein was extracted, and the NMR structure of the homologous protein of SARS-CoV (PDB code: 2MM4) was used as a template. The starting 3D model was then built using the Homology Modeling protocol Prime of the Schrodinger Suite [25]. According to the Ramachandran plot analysis for 58 residues, 93.1% lie in the most favored regions, 6.9% in the allowed regions, and none in the disallowed regions. The missing C and N terminal residues, not present in the chosen template, were finally added.
2.3. Molecular dynamic simulations of E protein
In order to get a more realistic model of the SARS-CoV-2 Envelope protein, the starting 3D homology model was further optimized using molecular dynamics simulations prepared via the CHARMM-GUI [26] server and performed using standard GROMACS tools [27]. At first, the protein was shortly simulated (10 ns, 300 K) in neutralized solution. It allowed to get conformations in which the transmembrane (TM), the C-terminus and the N-terminus domains were well suited for only embedding of the TM domain into a phospholipidic double layer membrane. The chosen conformation of the protein obtained this way was then oriented and inserted into a small POPC double layer (60 molecules per layer) surrounded by water (29,472 atoms). A 100 ns long MD simulation of the obtained system was performed as proof of stability.
2.4. In silico docking of SARS-CoV and SARS-CoV-2 Envelope C-terminus to PALS1
PALS1 structure for the in silico docking of the E protein C-terminus octapeptides, was obtained by the reported PALS1-CRB1 complex (PDB code: 4UU5). It was prepared for docking with the Maestro protein preparation wizard, using default parameters [28]. Neither side-chain atoms nor residues were missing in the protein in the neighbourhood of CRB1 peptide at 5 Å of the protein. A search grid was generated with Glide5 by selecting the 8 C-terminal residues of CRB1 (PPAMERLI) to define the binding pocket, thus including the entire binding site of the peptide–protein complex.
Then, the 8 C-terminal residues of each E protein were built, i.e. EGVPDLLV and SRVPDLLV for SARS-CoV and SARS-CoV-2, respectively, whose protonation state was assigned with PROPKA. Using the peptide-protein docking protocol of Glide [29] multiple conformers of the peptide were generated, docked on the protein and post-processed using MM-GBSA.
3. Results
3.1. Identification of genomic differences between SARS-CoV-2 and SARS-CoV E proteins
Multiple sequence alignments showed a quasi-perfect identity between all genomes of bats, pangolin, SARS-CoV and SARS-CoV-2 in the N-terminal and transmembrane regions of the E protein: only few synonymous mutations were identified in these two regions (Supplementary Table 2).
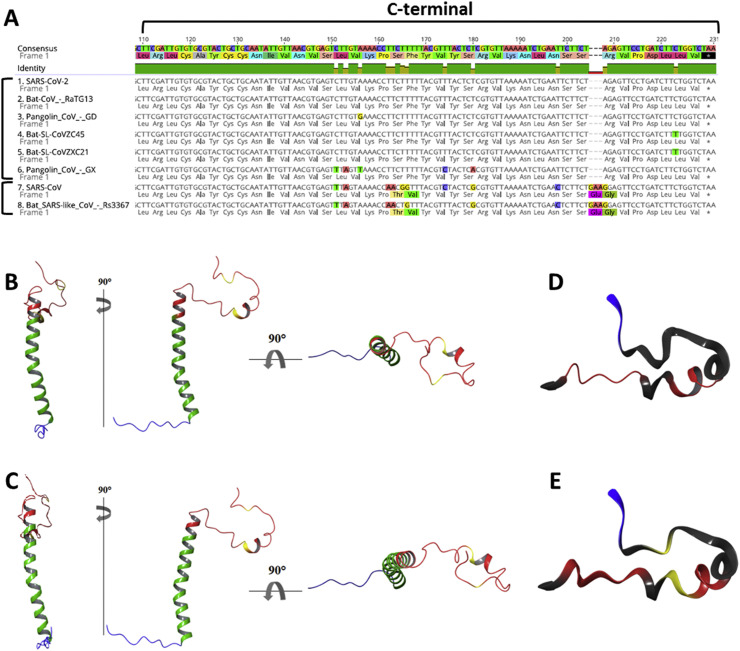
A different outcome is highlighted in the C-terminal region of the E protein sequence (Fig. 1 A) where two different mutation sites show a clear difference between SARS-CoV-2 and SARS-CoV, confirming what has been found from the whole genome phylogenetic analyses [2].
Fig. 1.
Multiple sequence alignment and homology modelling of E proteins. A) Envelope Protein multiple sequence alignment of C-terminal domain in human, bat and pangolin SARS-like coronaviruses highlights the identity among SARS-CoV-2, 2 bat CoV strains (RaTG13 and CoVZC45/CoVZX21) and pangolins E proteins. The comparison also points out two mutation sites where they differ from SARS-CoV and Bat SARS-like Cov Rs3367. B) E protein structure model for SARS and C) SARS-CoV-2. A closer look at C-terminal domain of E protein in D) SARS and E) SARS-CoV-2 in which are highlighted: in red, the motifs regulating the transport to Golgi apparatus; in blue, the PDZ-binding domain; in yellow, the two mutation sites identified in SARS-CoV-2 with respect to SARS.
Specifically, the mutation in the inner part of the C-terminal region consists in 4 nucleotide changes that lead to substitution of Thr55-Val56 for SARS-CoV with Ser55-Phe56 in SARS-CoV-2. The second mutation regards the deletion of Glu69-Gly70 and substitution with an Arg69, as also described by Hassan et al. [30].
3.2. Homology modelling of E proteins
In Fig. 1B, C, the two predicted monomeric E full length protein structure models have been constructed and show N-terminal (blue), transmembrane (green), C-terminal domains (red) as well as the amino acid variants (yellow). As expected, transmembrane domains of both proteins presented the highest accuracy with a total confidence score of more than 90% on ∼80% of the full-length proteins. The full-length domains of the SARS-CoV variants have been further characterised, posing them in a membrane bilayer and determining their stability on a short MD simulation of 100 ns (Supplementary Figure S1). In both models, as expected, the trans-membrane helix is perfectly fitted in the hydrophobic core of the membrane bilayer.
The end of the C-terminal, accounting 11 amino acid residues, and the beginning of the N-terminal end did not reach previously indicated accuracy. Moreover, deletion of two amino acid residues and arginine substitution at C-terminus could affect the protein structure altering the spatial disposition of the β-coil-β (Fig. 1D, E). The structure of this subunit appeared highly mobile but remained substantially unaltered along the short MD simulations for both E variants.
3.3. E protein C-terminals binding to tight junction-associated PALS1
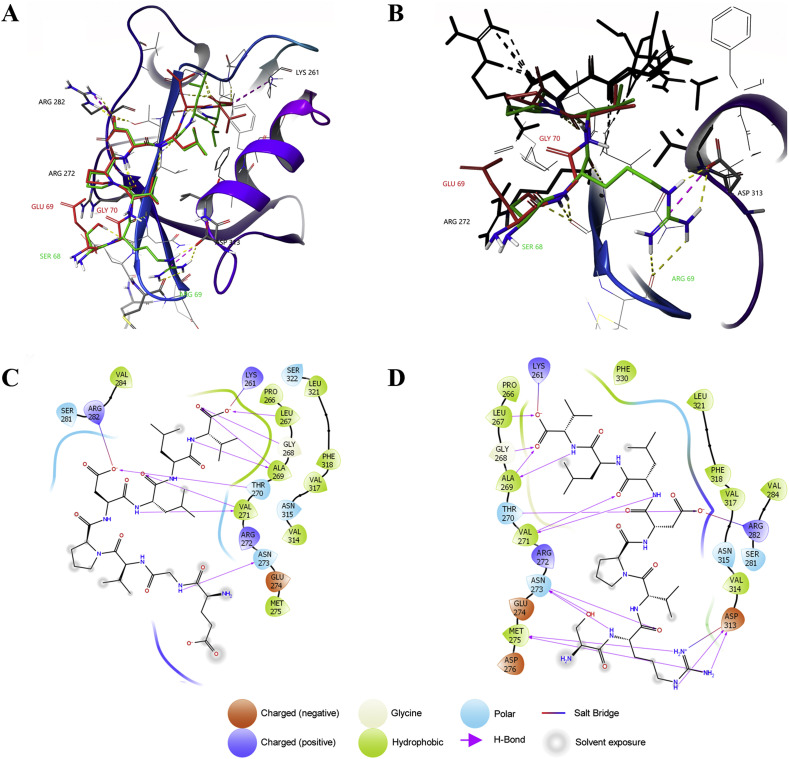
In order to verify the potential implications of the altered amino acid sequence, the binding pose of the two C-terminus octapeptides belonging to SARS-CoV and SARS-CoV-2 were determined and compared with the crystallographic structure of the complex PALS1-CRB1 [31]. The poses with the lowest ΔG, calculated via MM-GBSA by using default parameters, are shown in Fig. 2 A. Accordingly, the Free Energy of Binding for SARS-CoV and SARS-CoV-2 Envelope C-terminals amounts to −63.62 and −97.10 kcal/mol, respectively. This value must be compared to the value of −92.5 kcal/mol obtained performing the same analysis on the complex PALS1-CRB1, where the endogenous peptide was shortened to 8 amino acids in order to compare its in silico affinity with the SARS-CoVs variants. Interestingly, the SARS-CoV-2 peptide is able to bind PALS1 with a significantly higher affinity compared to SARS-CoV variant, reaching and slightly ameliorating the affinity value of the endogenous ligand, even though the two octapeptides differ for only two out of 8 of the selected amino acids. In particular, the last four residues of both E C-terminals are the same (Asp, Leu, Leu, Val) and bind PALS1 similarly to what observed for the endogenous CRB1, even if the short sequence of the CRB1 peptide is slightly different (Glu, Arg, Leu, Ile). As shown in the interaction maps described in Fig. 2C, D, the side-chain of the last residue of the E proteins, which is a valine, interacts with Leu267, Leu321 and Phe330 of PALS1; its free terminal carboxyl group, instead, makes a salt bridge with Lys261 and H-bond interactions with amide hydrogens of Leu267, Gly268 and Ala269. The two following leucine residues of the E C-terminals make van der Waals contacts, in particular the second one makes interaction with Phe318 (while CRB1 interacts with this residue via cation-π through its Arg). Aspartate is the last common residue inside the binding pocket and its sidechain residue makes a salt bridge with Arg282. Immediately after this negatively charged amino acid the two C-terminal residues of SARS-CoV and SARS-CoV-2 differ significantly. The positively charged Arg69 of the SARS-CoV-2 octapeptide well suits a negatively charged pocket (zoomed region in Fig. 2B), being able to contemporarily create a salt bridge with Asp313 and several H bonds with Asp313 and with a carbonyl oxygen of Met275 backbone. In the same position in SARS-CoV, the small size side chain of the Gly residue cannot be involved in any interaction with this pocket thus reducing the interaction strength. Finally, the Ser alcohol moiety of SARS-CoV-2 makes a hydrogen bond with Asp299 backbone oxygen, while the Glu residue in SARS-CoV could interact with Arg272 with a salt bridge but this condition is never realized among all the poses.
Fig. 2.
In silico docking of E proteins C-terminals with PALS1. A) SARS-CoV and SARS-CoV-2 octapeptides lowest ΔG poses on PALS1 binding site, representation of H bond in yellow dashed lines, Salt bridge in purple dashed lines (red structure and label: SARS-CoV; green structure and label: SARS-CoV-2). B) Magnification of SARS-CoV-2 Arg69 inside PALS1 negatively charged pocket showing the interesting salt bridge with ASP313 and H bonds with MET275. C) Ligand Interaction diagram of SARS-CoV octapeptide. D) Ligand Interaction diagram of SARS-CoV-2 octapeptide.
4. Discussion
In the present work, we compared the genomic sequences of SARS-CoV-2 E protein with the E protein of SARS-CoV, and the corresponding bats and pangolin orthologues, in order to identify the implications of dissimilarities in such an enigmatic protein.
Given the very small sequence length of the E protein, full genome multiple sequence alignment might affect the overall precision on this region, likely the reason for erroneously aligned E proteins in previous works, leading to misinterpret these amino acids deletion and substitution in the C-terminal end [32,33]. SARS-CoV matches to orthologue E proteins of bat-CoV Rs3367, while SARS-CoV-2 E protein is identical to bat RaTG13 and, except for synonymous mutations, to bat CoVs and the recently identified pangolin coronaviruses. Interestingly, SARS-CoV and SARS-CoV-2 E proteins confirm the two phylogenetic clades observed with the full genome [2].
The subsequent analysis aimed to identify potentially beneficial or detrimental effects of SARS-CoV-2 E protein variant with respect to the previously studied SARS E protein. Docking results of the tight junction complex PALS1-CRB1, used as a reference from the previous study by Ivanova and colleagues [31], were compared to the docking values of PALS1 with C-terminal ends of both E proteins. Our findings support the hypothesis that characteristic virulence of SARS-CoV-2 virus could depend on the strengthened interaction between SARS-CoV-2 E protein and PALS1 prompting a strong alteration of the tight junctions. The enhanced binding to PALS1 represents only the first step of the immunopathogenic process associated to SARS-CoV-2 infection. Moreover, PALS1 – E binding alters E-cadherin intracellular traffic with change in cell polarization [20] endorsing a severe dysregulation of the Th2 mediated response due to enhanced exposition to environmental allergens [34]. Epithelial mesenchymal transition occurs with a significant modification of the epithelial tissue and a meaningfully production of chemokines and cytokines in the infection site [35], promoting infection of nearby cells and potentially enhancing transmission to other individuals.
Remarkably, changes in epithelial tissue structures, and consequent loss of functions, are age-dependent and associated with defects in tight junctions and cadherin – catenin complex [36]. Although little information is available on lung epithelium ageing-dependent mechanisms, we hypothesize that E protein significantly increase this dysfunction, especially in elderly people as evidenced by COVID-19 epidemiological data. Conversely, SARS-CoV infection resulted in a major fatality rate compared with the recent SARS-CoV-2 infection. Further studies will be needed to elucidate the role of this genomic variant to promote novel interactions with PDZ domains in other host cell components.
Importance of our findings are corroborated by recent observations of the low rate of non-synonymous mutations in the genomic sequence codifying the E protein [30]. Indeed, these mutations have been detected only in ≈0.4% of the analysed genomes over the transmembrane and C-terminus domains. Interestingly, changes in the C-terminus motif observed in the QJR88103 (DLLV to DFLV) protein did not result in any alteration of the hydrophobicity, whereas in the QKI36831 protein the C-terminus YLLV showed a significant perturbation of the motif suggesting a diverse interaction with PALS1 [30]. Conservation of the E protein, and particularly of the sequence studied in this work, give therefore a key role to the C-terminal domain during infection.
The genomic variant highlighted in this work raise concerns on the underestimated role of the E protein of SARS-CoV-2 in the host adverse responses. Indeed, these computational results shed the lights on the most enigmatic protein among the structural proteins of coronaviruses, the E protein, laying the foundations for a fundamental detailed “wet” experimental investigation of its interaction with host components and comparison with E protein of SARS-CoV in standardized infection model.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
We would like to thank all scientists who have contributed sequences to the GISAID database (https://www.gisaid.org/). No funding was used to conduct this research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micinf.2020.08.006.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam T.T.-Y., Shum M.H.-H., Zhu H.-C., Tong Y.-G., Ni X.-B., Liao Y.-S. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 3.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong Y., Ren X. Coronavirus entry and release in polarized epithelial cells: a review. Rev Med Virol. 2014;24:308–315. doi: 10.1002/rmv.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sigrist C.J., Bridge A., Le Mercier P. A potential role for integrins in host cell entry by SARS-CoV-2. Antivir Res. 2020;177:104759. doi: 10.1016/j.antiviral.2020.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sims A.C., Burkett S.E., Yount B., Pickles R.J. SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium. Virus Res. 2008;133:33–44. doi: 10.1016/j.virusres.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.J Alsaadi E.A., Jones I.M. Membrane binding proteins of coronaviruses. Future Virol. 2019;14:275–286. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18:1–9. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kandeel M., Ibrahim A., Fayez M., Al-Nazawi M. From SARS and MERS CoVs to SARS-CoV-2: moving toward more biased codon usage in viral structural and nonstructural genes. J Med Virol. 2020;92:660–666. doi: 10.1002/jmv.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tilocca B., Soggiu A., Musella V., Britti D., Sanguinetti M., Urbani A. Molecular basis of COVID-19 relationships in different species: a one health perspective. Microb Infect. 2020;22:218–220. doi: 10.1016/j.micinf.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe Y., Allen J.D., Wrapp D., Mclellan J.S., Crispin M. Site-specific glycan analysis of the. SARS-CoV-2 spike. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tilocca B., Soggiu A., Sanguinetti M., Babini G., De Maio F., Britti D. Immunoinformatic analysis of the SARS-CoV-2 envelope protein as a strategy to assess cross-protection against COVID-19. Microb Infect. 2020;22:182–187. doi: 10.1016/j.micinf.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:1–22. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkatagopalan P., Daskalova S.M., Lopez L.A., Dolezal K.A., Hogue B.G. Coronavirus envelope (E) protein remains at the site of assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres J., Parthasarathy K., Lin X., Saravanan R., Kukol A., Ding X.L. Model of a putative pore: the pentameric α-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys J. 2006;91:938–947. doi: 10.1529/biophysj.105.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nieto-Torres J.L., DeDiego M.L., Álvarez E., Jiménez-Guardeño J.M., Regla-Nava J.A., Llorente M. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Surya W., Claudine S., Torres J. Structure of a conserved golgi complex-targeting signal in coronavirus envelope proteins. J Biol Chem. 2014;289:12535–12549. doi: 10.1074/jbc.M114.560094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen J.R., Lin L.D., Machamer C.E. Identification of a golgi complex-targeting signal in the cytoplasmic tail of the severe acute respiratory syndrome coronavirus Envelope protein. J Virol. 2011;85:5794–5803. doi: 10.1128/JVI.00060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teoh K.T., Siu Y.L., Chan W.L., Schlüter M.A., Liu C.J., Peiris J.S.M. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol Biol Cell. 2010;21:3838–3852. doi: 10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y., Wei Z., Yan Y., Wan Q., Du Q., Zhang M. Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex. Proc Natl Acad Sci U S A. 2014;111:17444–17449. doi: 10.1073/pnas.1416515111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh K. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson M.P., Pincus D.L., Rapp C.S., Day T.J.F., Honig B., Shaw D.E. A hierarchical approach to all-atom protein loop prediction. Proteins Struct Funct Genet. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 26.Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theor Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 28.Sastry M.G., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 29.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 30.Hassan S.S., Choudhury P.P., Roy B. SARS-CoV2 Envelope protein: non-synonymous mutations and its consequences. Genomics. 2020;112:3890–3892. doi: 10.1016/j.ygeno.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanova M.E., Fletcher G.C., O’Reilly N., Purkiss A.G., Thompson B.J., McDonald N.Q. Structures of the human Pals1 PDZ domain with and without ligand suggest gated access of Crb to the PDZ peptide-binding groove. Acta Crystallogr Sect D Biol Crystallogr. 2015;D71:555–564. doi: 10.1107/S139900471402776X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chatterjee S. Understanding the nature of variations in structural sequences coding for coronavirus spike, envelope, membrane and nucleocapsid proteins of SARS-CoV-2. PapersSsrnCom. 2020:1–12. [Google Scholar]
- 33.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P. Genome composition and divergence of the novel coronavirus ( 2019-nCoV ) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Georas S.N., Rezaee F. Epithelial barrier function: at the front line of asthma immunology and allergic airway inflammation. J Allergy Clin Immunol. 2014;134:509–520. doi: 10.1016/j.jaci.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartis D., Mise N., Mahida R.Y., Eickelberg O., Thickett D.R. Epithelial-mesenchymal transition in lung development and disease: does it exist and is it important? Thorax. 2014;69:760–765. doi: 10.1136/thoraxjnl-2013-204608. [DOI] [PubMed] [Google Scholar]
- 36.Parrish A.R. The impact of aging on epithelial barriers. Tissue Barriers. 2017;5 doi: 10.1080/21688370.2017.1343172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.