Abstract
Introduction
The rising burden of dementia is a global concern, and there is a need to study its causes, natural history and outcomes. The Secure Anonymised Information Linkage (SAIL) Databank contains anonymised, routinely-collected healthcare data for the population of Wales, UK. It has potential to be a valuable resource for dementia research owing to its size, long follow-up time and prospective collection of data during clinical care.
Objectives
We aimed to apply reproducible methods to create the SAIL dementia e-cohort (SAIL-DeC). We created SAIL-DeC with a view to maximising its utility for a broad range of research questions whilst minimising duplication of effort for researchers.
Methods
SAIL contains individual-level, linked primary care, hospital admission, mortality and demographic data. Data are currently available until 2018 and future updates will extend participant follow-up time. We included participants who were born between 1st January 1900 and 1st January 1958 and for whom primary care data were available. We applied algorithms consisting of International Classification of Diseases (versions 9 and 10) and Read (version 2) codes to identify participants with and without all-cause dementia and dementia subtypes. We also created derived variables for comorbidities and risk factors.
Results
From 4.4 million unique participants in SAIL, 1.2 million met the cohort inclusion criteria, resulting in 18.8 million person-years of follow-up. Of these, 129,650 (10%) developed all-cause dementia, with 77,978 (60%) having dementia subtype codes. Alzheimer’s disease was the most common subtype diagnosis (62%). Among the dementia cases, the median duration of observation time was 14 years.
Conclusion
We have created a generalisable, national dementia e-cohort, aimed at facilitating epidemiological dementia research.
Introduction
Dementia is a major global health challenge [1,2] and the current lack of disease-modifying therapies places the onus on the research community to identify potentially modifiable risk factors [3], as well as to study its incidence, prevalence and natural history. The pathologies underlying neurodegenerative diseases such as Alzheimer’s disease are likely to begin many years before symptom onset [4], and so long follow-up times are required to determine whether an association between a given factor and dementia is truly causal or due to reverse causation [5,6]. Longitudinal studies, with prospective data collection, are therefore of great importance to improving our understanding of dementia.
Dementias Platform UK (DPUK, www.dementiasplatform.uk) is a UK-wide, public-private partnership, that aims to facilitate and accelerate dementia research by providing a single point of access to data for >2 million participants across >38 existing cohort studies. Many of these cohorts have provided, and will continue to provide, important insights in the field of dementia. However, as these cohorts require participant consent for recruitment, they are likely to suffer from selection bias [7–9]. In contrast, a nationwide cohort based on whole-population administrative data is likely to avoid this issue, as analyses based on it can be more readily generalised to other populations.
The Secure Anonymised Information Linkage (SAIL) Databank (https://saildatabank.com) is a remotely-accessible, privacy-protecting data safe haven containing anonymised, individual-level, linked routinely-collected health and social care datasets for the population of Wales, UK [10–12]. Wales, with a population of approximately three million people, is one of four countries in the UK. A key enabler of data creation is Wales’ National Health Service (NHS), which acts a single provider of healthcare, free at the point of use to the resident population. As a result, SAIL is a large, nationwide, population-based research resource comprising longitudinal, routinely-collected healthcare data. Its size, national coverage and richness of available data means SAIL has the potential to be of great value for dementia research [13]. However, it can be a demanding task for researchers to transform the complex and varied datasets into a study population appropriate for their research question, as well as to identify participants with dementia with a minimum of misclassification.
By applying coding algorithms to linked routinely-collected datasets, we developed a novel DPUK cohort – the SAIL Dementia e-Cohort (SAIL-DeC). SAIL-Dec is a population-based electronic cohort (e-cohort) containing health-related information on people with and without diagnosed dementia. We developed SAIL-DeC to maximise its generalisability and utility for a broad range of research questions and methodologies. For example, we anticipate SAIL-DeC data being used to conduct risk factor studies, explore geographical variations in dementia incidence or outcomes, develop or validate risk prediction models and perform health economic analyses.
We created SAIL-DeC with the aims of minimising duplication of effort, increasing reproducibility, reducing costs and allowing a broader range of researchers to apply to use SAIL data.
Methods
Study reporting
We have followed the Reporting of studies Conducted using Observational Routinely-collected Data (RECORD) guideline in formatting this manuscript [14]. The SQL script used to create the cohort and cohort meta-data are available at https://datashare.is.ed.ac.uk/handle/10283/3268.
SAIL Databank
The SAIL Databank, based at Swansea University, was developed based on four principles: (1) to operate a remote access system, providing secure access to data to approved researchers; (2) to provide a powerful data analytic platform; (3) to ensure a robust mechanism for the safe transfer of approved files in and out of the system; and (4) to be efficient and scalable [11].
SAIL uses a split-file anonymisation method to maintain confidentiality. Individuals within each routinely-collected dataset are assigned a unique identifier (Anonymised Linking Field [ALF]). The ALF is generated by NHS Wales Information Service, a trusted third party, using the Matching Algorithm for Consistent Results in Anonymised Linkage, which has an accuracy of 99.85% [12,13]. Within SAIL, the ALF is further encrypted (ALF-E) and used to link the now de-identified individuals across multiple routinely-collected datasets, with further encryption (ALF_PE) then applied before data are allocated to an approved project.
Datasets
To construct SAIL-DeC, we used linked primary care, hospital admissions, mortality and deprivation datasets (https://saildatabank.com/saildata/sail-datasets). Hospital admissions data (Patient Episode Database for Wales [PEDW]), first collected in Wales in April 1991, contain information regarding inpatient admissions (emergency, elective and maternity) and day-case procedures. Diagnoses within PEDW are coded using the International Classification of Diseases version 10 (ICD-10) system [15]. Mortality data (Annual District Death Extract [ADDE]), available in SAIL since 1995 and derived from England and Wales’ death certification and registration system, contain diagnoses of cause of death as well as contributory comorbidities. ADDE uses ICD-9 coding until 2001 [16], and ICD-10 coding thereafter. Primary care data (Welsh Longitudinal General Practice dataset [WLGP]), currently use the Read version 2 system [17,18], although this will ultimately be replaced with the international SNOMED CT system in the future [19]. While ICD-10 codes contain only diagnostic information, Read codes contain information on diagnoses, administrative procedures, prescriptions, and symptoms and signs, making them a potentially rich resource for a wide range of research. Currently, SAIL contains primary care data for approximately 80% of the Welsh population. The subpopulation for whom primary care data are available are representative of the entire Welsh population in terms of age, sex and deprivation (Supplementary Appendix 1). The period of time covered by primary care data varies considerably between individuals and across practices – we included all available primary care data for all eligible participants. Deprivation data were derived from the Welsh Demographic Service Dataset (WDSD). Within WDSD, the Welsh Index of Multiple Deprivation [WIMD], is used to measure relative deprivation based on geographical household location, for small areas in Wales (Lower-layer Super Output Areas [LSOAs])[20]. To create SAIL-DeC we used the 2011 version of WIMD, linked to the 2001 version of LSOAs. In future updates of the cohort we will be able to update these deprivation datasets as new versions become available.
Study population
We included all participants within the SAIL Databank for whom primary care data were available, based on being registered with a SAIL-contributing general practice (GP) at any point. We excluded participants with a date of birth listed as before 1st January 1900, as we deemed these to be likely to be incorrect. We also excluded participants whose 60th birthday would fall after the latest date of follow-up, because the prevalence of dementia is very low below this age [21]. We therefore included participants born between 1st January 1900 and 1st January 1958 in the initial cohort development. The later date will change as the SAIL Databank receives updates of the datasets in the future, meaning the cohort will continue to increase in size over time as more participants become eligible. The timing of cohort refreshes will be negotiated with prospective applicants and available updates added to relevant extracts within SAIL-DeC.
We defined the entry date into the cohort as the first date of registration with a SAIL GP. We excluded participants without a valid GP registration date. We defined the last date of follow-up as the earliest of GP de-registration or death (currently January 2018).
Cohort tables
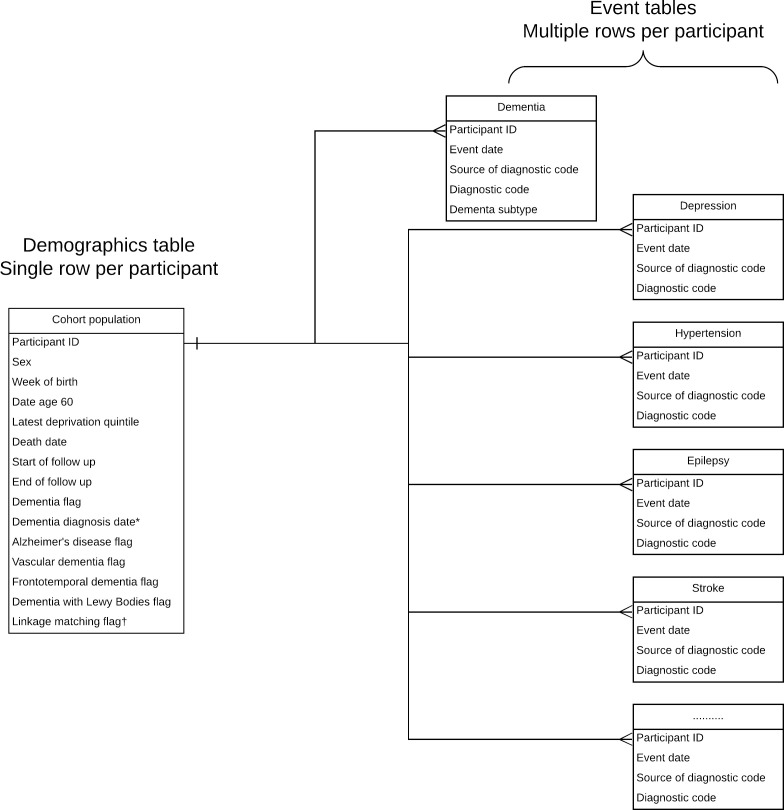
Using information from the four datasets, we created three types of table (Figure 1):
Figure 1: Format of SAIL dementia e-cohort.
Event tables consist of derived variables for: ever-smoking; obesity; hypertension; atrial fibrillation; peripheral arterial disease; myocardial infarction; stroke; diabetes; asthma; cancer; chronic obstructive pulmonary disease; depression; epilepsy; heart failure; hypothyroidism; osteoporosis; alcohol dependence; substance misuse; motor neurone disease; Parkinson’s disease; and rheumatoid arthritis.
*field blank if participant did not develop dementia.
†Flag indicating that linkage to ≥ 1 datasets for this participant may not be accurate (<95% probabilistic matching).
A demographics table, with one row per participant. This table holds basic demographic information and information on the follow-up time to allow survival analysis and/or efficient case-control matching. This table also contains information on the death date if appropriate and indicator flags on whether the participant developed all-cause dementia during follow-up. There is also a flag for whether the participant received a dementia subtype code.
A dementia events table, with multiple rows per participant. This contains the code details, date and source (i.e. primary care, hospital admissions or mortality data) of each dementia code and the dementia subtype to which the code refers.
Multiple risk factor/comorbidities events tables, with multiple rows per participant. Using the same format as the dementia events table, these tables contain information on the specific code, date and data source for each derived risk factor or comorbidity.
We created a data dictionary, which lists all cohort tables and outlines the source of the derived variables for each table type (Supplementary Appendix 2).
Derived variables
Dementia
We used a validated list of ICD-9, ICD-10 and Read V2 codes to identify all-cause dementia, Alzheimer’s disease, vascular dementia cases, dementia with Lewy Bodies and frontotemporal dementia in primary care, hospital admissions or mortality data (Supplementary Appendix 3). We developed the code list based on findings from a systematic review of the accuracy of dementia coding in routinely-collected healthcare data [22] and a UK-based validation study in which cases identified from coded data were compared to the full-text medical record [23]. We defined the date of diagnosis in the demographics table as the date of the first all-cause dementia code in any dataset. Where a participant had a dementia subtype code (e.g. Alzheimer’s disease), we defined date of diagnosis as the first date of any (all-cause) dementia code. Subtype code categories were not mutually exclusive, so a participant with ≥ 1 Alzheimer’s disease and vascular dementia codes in any dataset would be categorised as having both of these subtype diagnoses.
Risk factors and comorbidities
We created ICD-9, ICD-10 and Read V2 code lists to derive variables for risk factors and comorbidities based on a four-stage process: (1) code lists used by existing studies [24–34]; (2) an online clinical codes repository [35]; (3) where available, the recommended Read code lists from the UK Quality Outcomes Framework (QOF) [36] and (4) a manual review of the codes by a clinician (TW) (Supplementary Appendices 4-6). Where possible, we used validated code lists with known accuracy versus a definable reference standard; however, this was not possible for the majority of variables. Researchers who apply to use data from SAIL-DeC can use these code lists, or create their own using the underlying datasets, thereby creating an iterative process in which we use feedback from users of SAIL-DeC to improve and create different versions of the definitions of derived variables over time.
Bias
As the SAIL Databank contains data for the entirety of Wales, and primary care data for 80% of the population, we have attempted to minimise selection bias by including all eligible participants. We created code lists for diagnoses with the intention of maximising positive predictive value (PPV, the proportion of identified cases that are true cases) whilst maintaining a reasonable sensitivity (the proportion of true disease cases identified), in order to minimise bias in effect estimates [37].
Statistical methods
We calculated the total number of people and number of person-years of follow-up, stratified by sex, deprivation and birth decade for the whole cohort and for the dementia cases.
For the dementia cases, we counted the number of participants who had a specific dementia subtype code (Alzheimer’s disease, vascular dementia, dementia with Lewy Bodies and frontotemporal dementia). We calculated the median duration of follow-up for the dementia cases, as well as the number of cases and follow-up time for dementia cases in whom follow-up began prior to age 60. We also created an event flow diagram, indicating to what extent and in which order dementia cases were identified across multiple datasets. We calculated the number of dementia cases and person-years at risk for each derived risk factor or comorbidity.
Data access and cleaning methods
To create SAIL-DeC, we accessed all hospital admissions, mortality, primary care and deprivation data contained within SAIL. For the purposes of data cleaning, we excluded:
Participants with a recorded date of birth before 1/1/1900;
Any information from mortality data when the date of death was recorded as being before 1/1/1980 or after 1/1/2020. Where a record in mortality data was missing but death was recorded in the WDSD dataset, we used the latter to obtain the death date (7% of deceased participants);
Participants without a GP registration start or end date;
Participants who had a dementia diagnosis without a valid date (i.e. before 1/1/1900 or after 1/1/2018), as we did not know whether they represented true cases.
SAIL contains information on the linkage quality of ALFs obtained following deterministic and probabilistic matching which have been through a standard split file approach. We did not exclude participants based on low ALF matching rates, but instead created a flag in the demographics table to indicate where a participant has one or more linkages with <95% probabilistic matching. Users of SAIL-DeC can therefore opt to exclude participants of lower linkage quality depending on their study requirements.
Results
Demographics of whole cohort and dementia cases
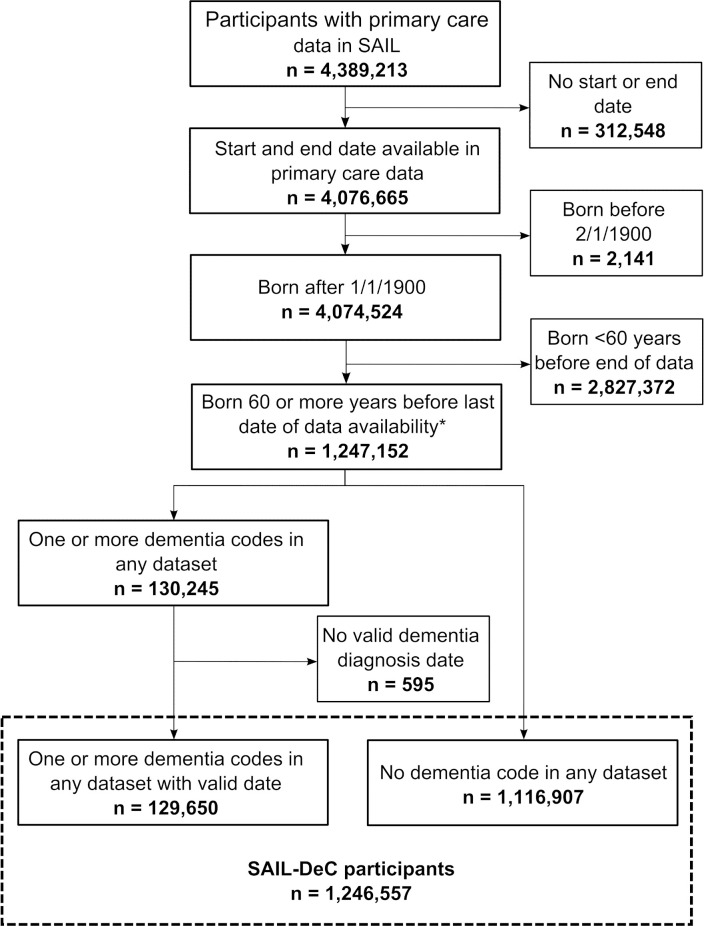
From the 4,389,213 people within the SAIL Databank with primary care data, 1,246,557 participants met the cohort inclusion criteria (Figure 2), resulting in 18,802,369 person-years of follow-up. For the whole cohort, the median first GP registration date was October 1995, with a median age at registration of 59 years. For participants with a diagnosis of dementia, the median date of first GP registration was January 1996 and median age at registration was 71 years. The demographics of the whole cohort and the dementia cases are displayed in Table 1. In the whole cohort, participants were equally distributed across deprivation quintiles.
Figure 2: Study flow diagram.
*Currently April 2016
*Latest deprivation score for each participant
Demographics of dementia cases and whole cohort.
| Whole cohort | Dementia cases | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| n | (%) | Person-years | n | (%) | Person-years | |
| Total participants | 1,246,557 | 18,796,117 | 129,650 | 1,773,462 | ||
| Sex | ||||||
| Female | 658,518 | (53) | 9,963,822 | 82,571 | (64) | 1,118,479 |
| Male | 588,035 | (47) | 8,832,289 | 47,079 | (36) | 654,983 |
| Missing | <5 | (0) | 5 | 0 | (0) | 0 |
| Deprivation quintile* | ||||||
| 1 (Most deprived) | 224,376 | (18) | 3,276,397 | 24,915 | (19) | 334,806 |
| 2 | 238,021 | (19) | 3,438,409 | 26,344 | (20) | 353,203 |
| 3 | 264,799 | (21) | 3,939,140 | 27,798 | (21) | 377,314 |
| 4 | 242,115 | (19) | 3,696,385 | 25,317 | (20) | 346,736 |
| 5 (Least deprived) | 255,363 | (20) | 4,223,548 | 24,033 | (19) | 350,798 |
| Missing | 21,883 | (2) | 222,238 | 1,243 | (1) | 10,604 |
| Birth Decade | ||||||
| 1900-1910 | 40,380 | (3) | 231,761 | 7,633 | (6) | 51,713 |
| 1911-1920 | 130,388 | (10) | 1,157,531 | 32,515 | (25) | 334,063 |
| 1921-1930 | 228,475 | (18) | 2,962,413 | 50,179 | (39) | 720,464 |
| 1931-1940 | 271,791 | (22) | 4,451,912 | 27,922 | (22) | 470,036 |
| 1941-1950 | 373,971 | (30) | 6,500,674 | 9,450 | (7) | 163,509 |
| 1951-1960 | 201,552 | (16) | 3,491,826 | 1,951 | (2) | 33,677 |
Dementia cases
Dementia subtypes
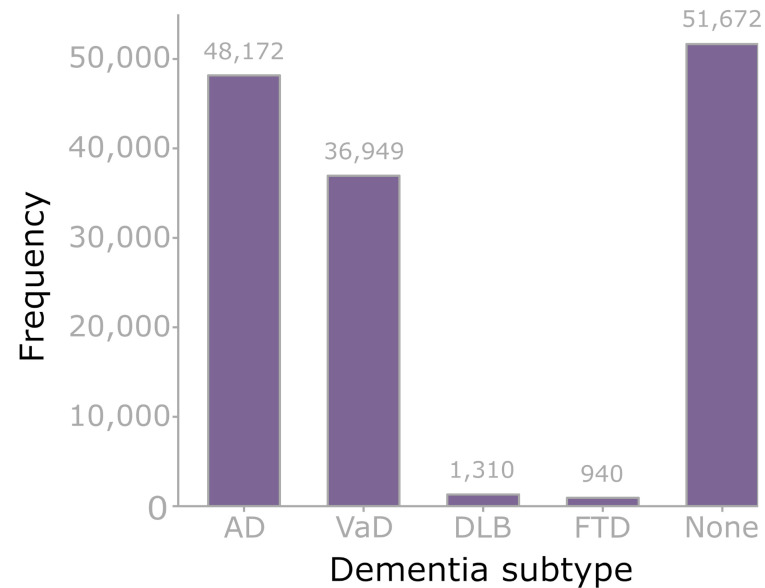
Of all SAIL-DeC participants, 129,650 (10%) developed all-cause dementia during follow-up. Of these, 77,978 (60%) had ≥ 1 codes for a dementia subtype (Figure 3). Alzheimer’s disease was the most common subtype diagnosis (48,172, 62%), followed by vascular dementia (36,949, 47%). 8,653 (11%) participants with dementia had both Alzheimer’s disease and vascular dementia codes.
Figure 3: Dementia subtypes.
AD – Alzheimer’s disease, VaD – vascular dementia, DLB – dementia with Lewy Bodies, FTD – frontotemporal dementia, None – no subtype code. Categories not mutually exclusive (apart from ‘none’ category). 77,978/129,650 (60.1%) of participants with all-cause dementia had at least one dementia subtype code. 8653 participants had both Alzheimer’s disease and vascular dementia codes (‘mixed dementia’).
Case ascertainment across datasets
From the 129,650 dementia cases, 78,828 (61%) were identified at any time in primary care data, 101,654 (78%) in hospital admissions data and 52,198 (40%) in mortality data. Forty-eight percent of participants were first identified in primary care data, 48% in hospital admissions data and 4% were identified only in mortality data (Supplementary Appendix 7). The number of dementia cases identified by each code are summarised in Supplementary Appendix 8.
Observation period
Among dementia cases, the median duration of follow-up was 14 years. We were able to follow up 23,724 (18%) dementia cases from <age 60 years, with a median follow-up time of 22 years. Seventy-nine percent of participants who developed dementia died during follow-up.
Risk factors and comorbidities
The number of participants and duration of follow-up for each risk factor or comorbidity among the 129,650 dementia cases is displayed in Table 2. A detailed breakdown of the number of participants identified by individual codes, as well as the extent to which participants with each risk factor or comorbidity are identified in the datasets over time, is available in Supplementary Appendices 9-29 and at https://datashare.is.ed.ac.uk/handle/10283/3268.
Table 2: Numbers and person-years of follow-up for risk factors or comorbidities across 129,650 dementia cases.
*Chronic kidney disease stages III-V. ICD-9, ICD-10 and Read V2 codes used to derive the variables are displayed in Supplementary Appendices 4-6. A detailed breakdown of the number of participants identified by each individual code is available in Supplementary Appendices 9-29 and at https://datashare.is.ed.ac.uk/handle/10283/3268.
| Risk factor / comorbidity | Dementia cases | ||
|---|---|---|---|
|
|
|||
| n | (%) | Person-years | |
| Ever-smoker | 67,775 | (52) | 1,070,030 |
| Obesity | 9,197 | (7) | 161,496 |
| Hypertension | 81,377 | (63) | 1,211,400 |
| Diabetes mellitus | 29,745 | (23) | 430,198 |
| Osteoporosis | 24,486 | (19) | 381,686 |
| Atrial fibrillation | 38,866 | (30) | 556,328 |
| Myocardial infarction | 22,194 | (17) | 310,403 |
| Heart failure | 31,703 | (24) | 426,072 |
| Peripheral arterial disease | 9,612 | (7) | 136,934 |
| Stroke | 32,240 | (25) | 427,974 |
| Depression | 39,130 | (30) | 587,340 |
| Parkinson’s disease | 9,845 | (8) | 128,771 |
| Epilepsy | 9,496 | (7) | 128,487 |
| Motor neurone disease | 332 | (0) | 4,284 |
| Asthma | 22,303 | (17) | 338,197 |
| Chronic obstructive pulmonary disease | 24,444 | (19) | 346,059 |
| Cancer | 34,663 | (27) | 496,564 |
| Rheumatoid arthritis | 5,597 | (4) | 82,091 |
| Hypothyroidism | 18,663 | (14) | 273,953 |
| Alcohol dependence | 6,289 | (5) | 89,684 |
| Substance misuse | 2,323 | (2) | 36,589 |
| Chronic kidney disease* | 26,821 | (21) | 447,895 |
Discussion
We have applied algorithms to routinely-collected primary care, hospital admissions, mortality and deprivation datasets within the SAIL Databank to create a ‘real world’ dementia e-cohort. We have incorporated this cohort into DPUK, to complement existing, ‘consented’ cohort studies within the initiative.
Flexibility
SAIL-DeC is designed to be flexible, meaning researchers can choose to use the existing disease definitions or create new ones to suit their purposes. For example, we used a validated code list for dementia outcomes in which the presence of a single dementia code in any dataset leads to a participant being identified as a dementia case, with the date of the first dementia code used to determine the ‘date of diagnosis’. Users may wish to use an alternative definition of dementia – for example, requiring >1 dementia codes in any data source, or including prescriptions for dementia drugs (e.g., cholinesterase inhibitors) in the algorithm. Similarly, they may wish to create new comorbidity variables or adapt the existing ones. We included all available data for all eligible participants, allowing users of SAIL-DeC to create a sub-cohort relevant to their research question. Researchers who intend to use SAIL-DeC as a cohort study (e.g. when conducting a case-cohort analysis) will need to select a time point from which follow up starts for an individual, such as a specific date or participant age.
Identifying dementia cases using routinely-collected healthcare data
For routinely-collected healthcare datasets to be used to identify dementia cases for research, they must do so with sufficient accuracy [22]. If using the data for analyses of risk factors or the natural history of dementia, identifying disease outcomes with a high PPV is important in order to minimise the risk of bias (37). Validation studies of UK routinely-collected healthcare data to identify dementia cases have reported PPVs of 83-100% [23,38,39], 85-87% [23,40] and 80-90% [23,41] for primary care, hospital admissions and mortality data respectively.
The sensitivity (the proportion of true disease cases identified) of using routinely-collected healthcare data to identify disease outcomes is another important consideration. There is a trade-off between PPV and sensitivity, meaning case ascertainment methods with a high PPV may fail to identify a proportion of ‘true’ cases. Studies of the sensitivity of hospital admissions and mortality data in patients known to mental health services with dementia (and therefore likely to overestimate sensitivity as it does not account for the proportion of people with dementia who are undiagnosed) reported estimates of 78% and 54% respectively [42,43]. The sensitivity of UK primary care data is currently unknown [22], although a study is underway to investigate this [44].
The use of multiple data sources improves our understanding of the timing of a dementia diagnosis, as in some cases there can be a significant delay between the identification of a participant with dementia in one dataset compared to others (Supplementary Appendix 7). This is particularly important for analyses in which the date of dementia diagnosis is needed, such as time-to-event analyses.
Potential uses
There are numerous potential uses for this cohort. Given the breadth of primary care Read codes, there is the opportunity to identify novel risk factors for dementia and its subtypes. For example, there is increasing evidence that some drugs are associated with an increased risk of dementia [45–47]. The primary care dataset within SAIL-DeC contains details of all drug prescriptions, meaning the cohort could be used to explore this issue. Recent work by the Whitehall II study has shown the importance of long follow-up times to explore whether associations between various factors and dementia are in fact due to the effects of dementia on the factor itself (reverse causation) [5,6]. The long follow-up times in SAIL-DeC would enable such studies for a variety of risk factors.
Routinely-collected healthcare datasets have been used to study dementia incidence and prevalence [48,49], as well as to investigate within-country geographical variations in dementia outcomes [50]. As SAIL have obtained primary care data for a large proportion of the Welsh population, with included participants representative of the wider population, the dataset provides an opportunity to explore geographical variations in dementia incidence or outcomes.
Of the participants who developed dementia, 79% died during follow-up. This shows that, for many participants, follow-up until death was ‘completed’ (i.e. not censored early). SAIL-DeC would therefore be well suited to studies surrounding end of life for people diagnosed with dementia.
Primary care data have been used to develop and validate risk prediction models for a range of diseases [51–53]. Given that the variables within SAIL-DeC are all routinely-collected, any variables used in a dementia risk prediction model developed using the cohort’s data would be applicable to current clinical use.
The breadth of information contained within Read codes raises the possibility of using data within the cohort for hypothesis-free studies of dementia, such as environment-wide association studies [54,55]. With >100,000 dementia outcomes within SAIL-Dec, the cohort is likely to provide sufficient statistical power for such analyses.
In addition to hospital admissions data, SAIL also contains other healthcare datasets such as emergency attendances, critical care admissions, care home residence, pathology results and outpatient referrals. Other routine datasets are continuing to be linked to SAIL participants, and these could be linked to SAIL-DeC participants when they are made available. These datasets may provide a useful means with which to conduct health economic analyses for dementia care. There are also plans to derive new phenotype data from multiple sources of NHS clinical data using natural language processing, such as radiology reports and free-text correspondence between clinicians.
Accessing the data
Researchers interested in using SAIL-DeC data can contact the SAIL Databank directly (https://saildatabank.com/application-process), or approach DPUK via the DPUK portal (https://portal.dementiasplatform.uk), who can facilitate the application. Applicants must submit their proposal to the independent Information Governance Review Panel (IGRP), which ensures proper and appropriate use of SAIL data. Researchers are required to demonstrate appropriate Information Governance training prior to being provided with remote access to the SAIL safe haven [11]. Costs depend on the complexity of the project and support required and are outlined early in the project scoping process. Our intention is that using the pre-prepared SAIL-DeC datasets should reduce the complexity of data preparation, thereby minimising costs and time needed for new studies.
Strengths and limitations
SAIL-DeC has several strengths as a research resource. It is population-based, and contains data for ~80% of Wales, meaning that it is likely to be generalisable to other similar populations and should not suffer from the ‘healthy cohort effect’ [7–9]. This is reflected in the near-equal distribution of participants across deprivation quintiles. Its size is another strength. With 18.8 million person-years of follow-up in the whole cohort, there is likely to be sufficient power for most types of analyses, even on relatively rare exposures or outcomes. In creating the e-cohort, we have attempted to make it useful for a wide range of research studies, with the aim of increasing efficiency and reducing costs for researchers. We have used coded algorithms to simplify the routinely-collected datasets, with the intention of enabling researchers without experience of using UK healthcare datasets to use SAIL data. We have created derived variables for many comorbidities and risk factors, but the resource is designed to be flexible: users can request additional variables or alter how variables are defined if required for their analyses. Furthermore, if users change how some derived variables are defined (i.e. codes removed or added, or more complex algorithms created) based on their experience or the latest evidence, we can alter the definitions of these variables for other researchers too, creating a ‘learning’ resource for the dementia research community.
The e-cohort also has several limitations. First, routinely-collected healthcare data will not identify dementia cases with perfect accuracy. Whereas validation studies have shown PPV to be generally high across UK datasets, no studies have calculated the sensitivity of using primary care, hospital admissions and mortality data in combination [22]. To have the opportunity to appear in routinely-collected data, people with dementia must first be known to healthcare services with a dementia diagnosis, and dementia is known to be underdiagnosed [56], meaning sensitivity is likely to be lower than PPV. This means that SAIL-DeC would probably not be an appropriate resource with which to calculate absolute dementia prevalence and incidence in Wales as it will likely underestimate the number of cases. However, it could be used to compare the relative burden of dementia across different geographical areas.
The accuracy of the algorithms used to derive many of the variables for risk factors and comorbidities is not known. We have attempted to use algorithms with a presumed high PPV over sensitivity, by including codes we consider likely to reflect true positive cases and excluding codes that may introduce false positive cases. This means that some of our algorithms may identify risk factors and comorbidities with a low sensitivity, as suggested by the low rates of obesity (7%) at any point for the dementia cases. It is likely that some of the factors, particularly those for which recording is mandatory in QOF, are better recorded and therefore more likely to be detected than others. Our intention is that this will improve over time: as new validation studies of these variables are performed, and users of SAIL-DeC provide feedback on the code lists for these variables, we will update these algorithms to maximise their accuracy. For example, there is the potential to create more complex algorithms (e.g. by including continuous measurements such as body mass index for obesity or blood pressure readings for hypertension), to improve case ascertainment for some variables.
Although UK routinely-collected healthcare datasets identify all-cause dementia cases with a high PPV, the PPVs for the identification of dementia subtypes is lower. Using UK hospital admissions, mortality and primary care data in combination, PPVs were estimated as 71% for Alzheimer’s disease and 44% for vascular dementia [23]. There have been no validation studies estimating the PPVs for rare dementia subtypes such as dementia with Lewy Bodies and frontotemporal dementia [22]. Researchers should consider this when using SAIL-DeC to study dementia subtypes rather than all-cause dementia.
Furthermore, SAIL-DeC relies entirely on routinely-collected data to identify dementia cases as well as risk factors and other comorbidities. Although Read coding in primary care data provides a wide range of information in addition to diagnoses such as symptoms, signs, administrative procedures and prescriptions, there is limited phenotypic depth – for example there is no imaging, free-text or genetic data. Over time this may change, as SAIL obtains linkage to more detailed datasets.
The availability of primary care data in SAIL increased over time, as practices switched to electronic records, meaning the primary care records further back in time are less complete. We therefore have less information on participants earlier in life compared to later life, which may limit the use of cohort to study early or midlife risk factors for dementia. The introduction of QOF from 2004 onwards changed the way in which GPs were remunerated, and this led to changes in how GPs coded diagnoses and symptoms for certain conditions [57,58]. Dementia was introduced to QOF in 2006/2007, resulting in a sudden increase in dementia primary care codes around this time. Researchers using SAIL-DeC for survival analyses may wish to consider this when selecting their study time window.
Conclusion
In conclusion, we have applied coding algorithms to primary care, hospital admissions and mortality data to create SAIL-DeC, a national dementia e-cohort, to complement existing cohorts within DPUK. The cohort will enable researchers to conduct a wide range of analyses related to dementia, whilst minimising duplication of effort, time and cost.
Ethics statement
Ethical approval was not required because the study used only anonymised data. Approval was granted by the Information Governance Review Panel (IGRP, application number: 0697). Composed of government, regulatory and professional agencies, the IGRP oversees and approves applications to use the SAIL databank.
Supplementary Material
Acknowledgements
This study makes use of the Secure Anonymised Information Linkage (SAIL) Databank. We would like to acknowledge all the data providers who make anonymised data available for research. This work was supported by Health Data Research UK, which receives its funding from HDR UK Ltd (NIWA1) funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation (BHF) and the Wellcome Trust. We are grateful to Dementias Platform UK for supporting the development and creation of the SAIL dementia electronic cohort. We would like to acknowledge Kirsty Wilson, John Nolan, Qiuli Zhang, Kristiina Rannikmäe, Claire Tochel and Kathryn Bush for their advice on creating code lists for deriving comorbidity variables.
Abbreviations
| ADDE | Annual District Death Extract |
| ALF | Anonymised Linking Field |
| DPUK | Dementias Platform UK |
| GP | General practice |
| ICD | International Classification of Diseases |
| IGRP | Information Governance Review Panel |
| LSOA | Lower Super Output Area |
| NHS | National Health Service |
| PEDW | Patient Episode Database for Wales |
| PPV | Positive Predictive Value |
| QOF | Quality Outcomes Framework |
| Read V2 | Read version 2 |
| RECORD | The REporting of studies Conducted using Observational Routinely-collected health Data statement |
| SAIL | Secure Anonymised Information Linkage (Databank) |
| SAIL-DeC | SAIL dementia electronic cohort |
| SNOMED CT | Systematised Nomenclature of Medicine & Clinical Terms |
| UK | United Kingdom |
| WDSD | Welsh Demographic Service Dataset |
| WIMD | Welsh Index of Multiple Deprivation |
| WLGP | Welsh Longitudinal General Practice dataset |
Funding Statement
This work was funded by Dementias Platform UK (MR/L023784/2). TW is funded by a Medical Research Council Clinical Research Training Fellowship (MR/P001823/1).
References
- Prince M, Wimo A, Guerchet M, Ali G-C, Wu Y-T, Prina M. World Alzheimer Report 2015: The global impact of dementia. August2015.
- Wimo A, Guerchet M, Ali G-C, Wu Y-T, Prina AM, Winblad B, et al. The worldwide costs of dementia 2015 and comparisons with 2010. Alzheimers Dement. January2017;13(1):1-7. 10.1016/j.jalz.2016.07.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett J, Bird C, Ballard C, Banerjee S, Brayne C, Cowan K, et al. A roadmap to advance dementia research in prevention, diagnosis, intervention, and care by 2025. Int J Geriatr Psychiatry. July2018;33(7):900-906. 10.1002/gps.4868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman RJ, Xiong C, Benzinger TLS, Fagan AM, Goate A, Fox NC, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med. 30August2012;367(9):795-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivimäki M, Luukkonen R, Batty GD, Ferrie JE, Pentti J, Nyberg ST, et al. Body mass index and risk of dementia: Analysis of individual-level data from 1.3 million individuals. Alzheimers Dement. May2018;14(5):601-609. 10.1016/j.jalz.2017.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabia S, Dugravot A, Dartigues J-F, Abell J, Elbaz A, Kivimäki M, et al. Physical activity, cognitive decline, and risk of dementia: 28 year follow-up of Whitehall II cohort study. BMJ. 22June2017;357:j2709 10.1136/bmj.j2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon T, Moore FE, Shurtleff D, Dawber TR. Some methodologic problems in the long-term study of cardiovascular disease: Observations on the Framingham study. Journal of Chronic Diseases. 1September1959;10(3):186-206. [Google Scholar]
- Froom P, Melamed S, Kristal-Boneh E, Benbassat J, Ribak J. Healthy Volunteer Effect in Industrial Workers. Journal of Clinical Epidemiology. 1August1999;52(8):731-735. 10.1016/S0895-4356(99)00070-0 [DOI] [PubMed] [Google Scholar]
- Criqui MH, Austin M, Barrett-Connor E. The effect of non-response on risk ratios in a cardiovascular disease study. Journal of Chronic Diseases. 1January1979;32(9):633-638. 10.1016/0021-9681(79)90093-6 [DOI] [PubMed] [Google Scholar]
- Ford DV, Jones KH, Verplancke J-P, Lyons RA, John G, Brown G, et al. The SAIL Databank: building a national architecture for e-health research and evaluation. BMC Health Serv Res. 2009;9:157 10.1186/1472-6963-9-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KH, Ford DV, Jones C, Dsilva R, Thompson S, Brooks CJ, et al. A case study of the Secure Anonymous Information Linkage (SAIL) Gateway: A privacy-protecting remote access system for health-related research and evaluation. J Biomed Inform. August2014;50(100):196-204. 10.1016/j.jbi.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons RA, Jones KH, John G, Brooks CJ, Verplancke J-P, Ford DV, et al. The SAIL databank: linking multiple health and social care datasets. BMC Med Inform Decis Mak. 2009 Jan 16;9:3. 10.1186/1472-6947-9-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis M, Shine L, John A, Marchant A, McGregor J, Lyons RA, et al. Risk of Adverse Outcomes for Older People with Dementia Prescribed Antipsychotic Medication: A Population Based e-Cohort Study. Neurol Ther. 4January2017;6(1):57-77. 10.1007/s40120-016-0060-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchimol EI, Smeeth L, Guttmann A, Harron K, Moher D, Petersen I, et al. The REporting of studies Conducted using Observational Routinely-collected health Data (RECORD) Statement. PLOS Med. 6October2015;12(10):e1001885 10.1371/journal.pmed.1001885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. ICD-10 Version: 2016 [Internet]. 2016 Available from: http://apps.who.int/classifications/icd10/browse/2016/en
- World Health Organization. International Classification of Diseases: Ninth revision. Geneva; 1978. [Google Scholar]
- NHS Digital. Read Codes [Internet]. [cited 2017 May 17]. Available from: https://digital.nhs.uk/article/1104/Read-Codes
- Booth N. What are the Read Codes? Health Libr Rev. September1994;11(3):177-182. [DOI] [PubMed] [Google Scholar]
- SNOMED International. SNOMED CT [Internet]. [cited 2017 May 18]. Available from: http://www.snomed.org/snomed-ct
- Welsh Government. Welsh Index of Multiple Deprivation [Internet]. [cited 2018 Aug 23]. Available from: https://gov.wales/statistics-and-research/welsh-index-multiple-deprivation/?lang=en
- Alexander M, Perera G, Ford L, Arrighi HM, Foskett N, Debove C, et al. Age-Stratified Prevalence of Mild Cognitive Impairment and Dementia in European Populations: A Systematic Review. J Alzheimers Dis. 2015;48(2):355-359. 10.3233/JAD-150168 [DOI] [PubMed] [Google Scholar]
- Wilkinson T, Ly A, Schnier C, Rannikmäe K, Bush K, Brayne C, et al. Identifying dementia cases with routinely collected health data: A systematic review. Alzheimers Dement. August2018;14(8):1038-1051. 10.1016/j.jalz.2018.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson T, Schnier C, Bush K, Rannikmäe K, Henshall DE, Lerpiniere C, et al. Identifying dementia outcomes in UK Biobank: a validation study of primary care, hospital admissions and mortality data. Eur J Epidemiol [Internet]. 26February2019 [cited 2019 Feb 27]; Available from: 10.1007/s10654-019-00499-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A, Ashcroft DM, Owens L, van Staa TP, Pirmohamed M. Drug therapy for alcohol dependence in primary care in the UK: A Clinical Practice Research Datalink study. PLoS ONE. 2017;12(3):e0173272 10.1371/journal.pone.0173272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr MJ, Ashcroft DM, Kontopantelis E, While D, Awenat Y, Cooper J, et al. Premature Death Among Primary Care Patients With a History of Self-Harm. Ann Fam Med. 2017;15(3):246-254. 10.1370/afm.2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C, Webb RT, Carr MJ, Kontopantelis E, Green J, Chew-Graham CA, et al. Incidence, clinical management, and mortality risk following self harm among children and adolescents: cohort study in primary care. BMJ. 18October2017;359:j4351 10.1136/bmj.j4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorton HC, Webb RT, Carr MJ, DelPozo-Banos M, John A, Ashcroft DM. Risk of Unnatural Mortality in People With Epilepsy. JAMA Neurol. 1August2018;75(8):929-938. 10.1001/jamaneurol.2018.0333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran T, Kontopantelis E, Valderas JM, Campbell S, Roland M, Salisbury C, et al. Effect of financial incentives on incentivised and non-incentivised clinical activities: longitudinal analysis of data from the UK Quality and Outcomes Framework. BMJ. 28June2011;342:d3590 10.1136/bmj.d3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves D, Springate DA, Ashcroft DM, Ryan R, Doran T, Morris R, et al. Can analyses of electronic patient records be independently and externally validated? The effect of statins on the mortality of patients with ischaemic heart disease: a cohort study with nested case-control analysis. BMJ Open. 23April2014;4(4):e004952 10.1136/bmjopen-2014-004952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhurst C, Watt I, Martin F, Bland M, Brackenbury WJ. Exposure to sodium channel-inhibiting drugs and cancer survival: protocol for a cohort study using the QResearch primary care database. BMJ Open. 14November2014;4(11):e006604 10.1136/bmjopen-2014-006604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson A, Ford E, Davies KA, Smith HE, Rait G, Tate AR, et al. Optimising use of electronic health records to describe the presentation of rheumatoid arthritis in primary care: a strategy for developing code lists. PLoS ONE. 2013;8(2):e54878 10.1371/journal.pone.0054878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks SJ, Kontopantelis E, Akbarov A, Rodgers S, Avery AJ, Ashcroft DM. Examining variations in prescribing safety in UK general practice: cross sectional study using the Clinical Practice Research Datalink. BMJ. 3November2015;351:h5501 10.1136/bmj.h5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhurst C, Martin F, Watt I, Doran T, Bland M, Brackenbury WJ. Sodium channel-inhibiting drugs and cancer survival: protocol for a cohort study using the CPRD primary care database. BMJ Open. June2016;6(9):e011661 10.1136/bmjopen-2016-011661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springate DA, Ashcroft DM, Kontopantelis E, Doran T, Ryan R, Reeves D. Can analyses of electronic patient records be independently and externally validated? Study 2--the effect of β-adrenoceptor blocker therapy on cancer survival: a retrospective cohort study. BMJ Open. 13April2015;5(4):e007299 10.1136/bmjopen-2014-007299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springate DA, Kontopantelis E, Ashcroft DM, Olier I, Parisi R, Chamapiwa E, et al. ClinicalCodes: An Online Clinical Codes Repository to Improve the ValiKontopantelisdity and Reproducibility of Research Using Electronic Medical Records. PLOS ONE. 18June2014;9(6):e99825 10.1371/journal.pone.0099825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NHS Digital. QOF business rules v36.0 [Internet]. 2017 [cited 2017 Aug 30]. Available from: content.digital.nhs.uk/qofbrv36
- Chubak J, Pocobelli G, Weiss NS. Tradeoffs between accuracy measures for electronic health care data algorithms. J Clin Epidemiol. March2012;65(3):343-349.e2. 10.1016/j.jclinepi.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn N, Mullee M, Perry VH, Holmes C. Association between dementia and infectious disease: evidence from a case-control study. Alzheimer Dis Assoc Disord. June2005;19(2):91-94. 10.1097/01.wad.0000165511.52746.1f [DOI] [PubMed] [Google Scholar]
- Heath CA, Mercer SW, Guthrie B. Vascular comorbidities in younger people with dementia: a cross-sectional population-based study of 616 245 middle-aged people in Scotland. J Neurol Neurosurg Psychiatr. September2015;86(9):959-964. 10.1136/jnnp-2014-309033 [DOI] [PubMed] [Google Scholar]
- Brown A, Kirichek O, Balkwill A, Reeves G, Beral V, Sudlow C, et al. Comparison of dementia recorded in routinely collected hospital admission data in England with dementia recorded in primary care. Emerg Themes Epidemiol. 2016;13:11 10.1186/s12982-016-0053-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Calloway R, Zhao E, Brayne C, Matthews FE. Accuracy of death certification of dementia in population-based samples of older people: analysis over time. Age Ageing. 1July2018;47(4):589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerlad A, Perera G, Singh-Manoux A, Lewis G, Stewart R, Livingston G. Accuracy of general hospital dementia diagnoses in England: Sensitivity, specificity, and predictors of diagnostic accuracy 2008–2016. Alzheimer’s & Dementia. 25April2018;14(7):933-943. 10.1016/j.jalz.2018.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera G, Stewart R, Higginson IJ, Sleeman KE. Reporting of clinically diagnosed dementia on death certificates: retrospective cohort study. Age Ageing. 2016;45(5):668-673. 10.1093/ageing/afw077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldus C, Brayne C, Matthews F, Arthur A, Dening T, Fox C, et al. The prevalence, causes and consequences of undiagnosed dementia in England: a record linkage study of the Cognitive Function and Ageing Study II. Alzheimer’s & Dementia: The Journal of the Alzheimer’s Association. 1July2017;13(7):P1041 10.1016/j.jalz.2017.06.1470 [DOI] [Google Scholar]
- Coupland CAC, Hill T, Dening T, Morriss R, Moore M, Hippisley-Cox J. Anticholinergic Drug Exposure and the Risk of Dementia: A Nested Case-Control Study. JAMA Intern Med. 24June2019; 10.1001/jamainternmed.2019.0677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson K, Fox C, Maidment I, Steel N, Loke YK, Arthur A, et al. Anticholinergic drugs and risk of dementia: case-control study. BMJ. 25April2018;361:k1315 10.1136/bmj.k1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale H, Gomm W, Broich K, Maier W, Tolppanen A-M, Tanskanen A, et al. Use of Antiepileptic Drugs and Dementia Risk-an Analysis of Finnish Health Register and German Health Insurance Data. J Am Geriatr Soc. 22March2018; 10.1111/jgs.15358 [DOI] [PubMed] [Google Scholar]
- Perera G, Pedersen L, Ansel D, Alexander M, Arrighi HM, Avillach P, et al. Dementia prevalence and incidence in a federation of European Electronic Health Record databases: The European Medical Informatics Framework resource. Alzheimer’s & Dementia. 1February2018;14(2):130-139. 10.1016/j.jalz.2017.06.2270 [DOI] [PubMed] [Google Scholar]
- Pujades-Rodriguez M, Assi V, Gonzalez-Izquierdo A, Wilkinson T, Schnier C, Sudlow C, et al. The diagnosis, burden and prognosis of dementia: A record-linkage cohort study in England. PLOS ONE. 26June2018;13(6):e0199026 10.1371/journal.pone.0199026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ TC, Murianni L, Icaza G, Slachevsky A, Starr JM. Geographical Variation in Dementia Mortality in Italy, New Zealand, and Chile: The Impact of Latitude, Vitamin D, and Air Pollution. DEM. 2016;42(1–2):31-41. 10.1371/journal.pone.0199026 [DOI] [PubMed] [Google Scholar]
- Hippisley-Cox J, Coupland C, Brindle P. Development and validation of QRISK3 risk prediction algorithms to estimate future risk of cardiovascular disease: prospective cohort study. BMJ. 23May2017;357:j2099 10.1136/bmj.j2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters K, Hardoon S, Petersen I, Iliffe S, Omar RZ, Nazareth I, et al. Predicting dementia risk in primary care: development and validation of the Dementia Risk Score using routinely collected data. BMC Med. 21January2016;14:6. 10.1186/s12916-016-0549-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng SF, Reps J, Kai J, Garibaldi JM, Qureshi N. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS ONE. 2017;12(4):e0174944 10.1371/journal.pone.0174944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis DP, Brownstein JS, Patel CJ. Environment-Wide Association Study of Blood Pressure in the National Health and Nutrition Examination Survey (1999-2012). Sci Rep. 2016 26;6:30373 10.1038/srep30373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CJ, Bhattacharya J, Butte AJ. An Environment-Wide Association Study (EWAS) on type 2 diabetes mellitus. PLoS ONE. 20May2010;5(5):e10746 10.1371/journal.pone.0010746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang L, Clifford A, Wei L, Zhang D, Leung D, Augustine G, et al. Prevalence and determinants of undetected dementia in the community: a systematic literature review and a meta-analysis. BMJ Open. 1February2017;7(2):e011146 10.1136/bmjopen-2016-011146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick T, Stuart B, Newell C, Geraghty AWA, Moore M. Changes in rates of recorded depression in English primary care 2003-2013: Time trend analyses of effects of the economic recession, and the GP contract quality outcomes framework (QOF). J Affect Disord. 15July2015;180:68–78. 10.1016/j.jad.2015.03.040 [DOI] [PubMed] [Google Scholar]
- Tate AR, Dungey S, Glew S, Beloff N, Williams R, Williams T. Quality of recording of diabetes in the UK: how does the GP’s method of coding clinical data affect incidence estimates? Cross-sectional study using the CPRD database. BMJ Open. 1January2017;7(1):e012905 10.1136/bmjopen-2016-012905 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.