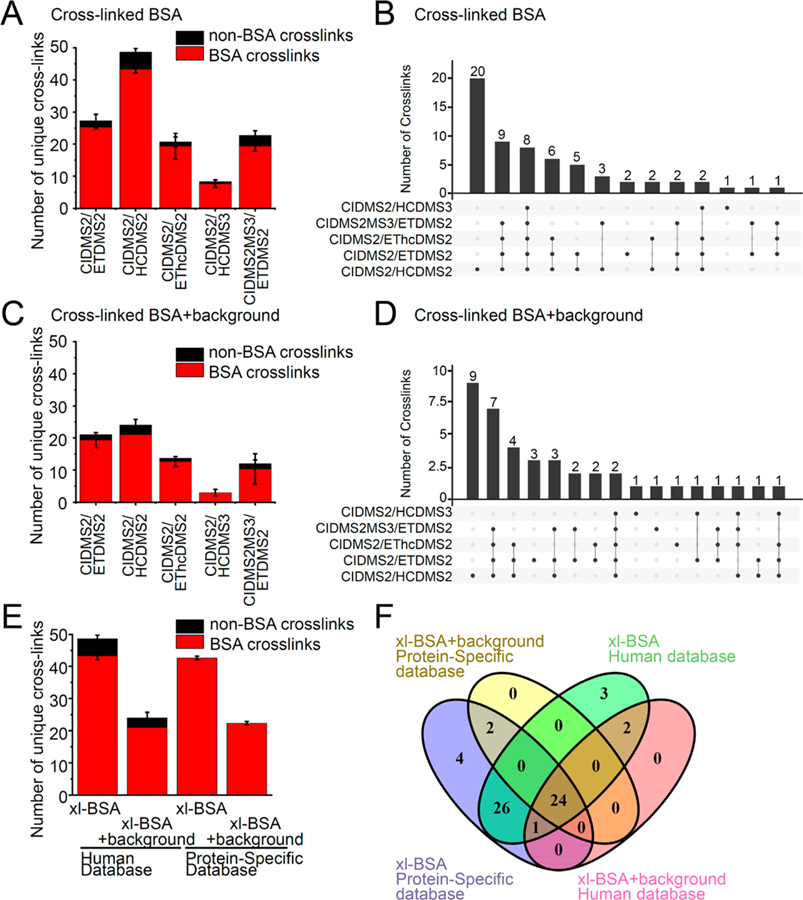
Figure 2.
Improved cross-linked peptide identification using CID-MS2/HCD-MS2 fragmentation. Comparison of 4 different fragmentation methods: CID-MS2/HCD-MS2, CID-MS2/ETD-MS2, CID-MS2/EThcD-MS2, CID-MS2/HCD-MS3, and CID-MS2-MS3/ETD-MS2 and databases: BSA protein specific database or human proteome database with BSA for increasing cross-link peptide identification. (A) Comparison between 5 fragmentation methods for average number of cross-link identifications of cross-linked BSA sample. BSA cross-links colored red and non-BSA cross-links colored black. Error bars represent standard deviation from three biological replicates. (B) Comparison between 5 fragmentation methods for overlap of all unique cross-link identifications. (C) Comparison between 5 fragmentation methods for average number of cross-link identifications of cross-linked BSA peptides spiked into human proteome background. (D) Comparison between 5 fragmentation methods for overlap of all unique cross-link identifications of cross-linked BSA sample spiked into human proteome background. (E) Comparison between 2 databases for average number of cross-link identifications of cross-linked BSA peptides and cross-linked BSA peptides spiked into human proteome background. (F) Comparison between 2 databases for overlap of all unique cross-link identifications of cross-linked BSA peptides and cross-linked BSA peptides spiked into human proteome background.