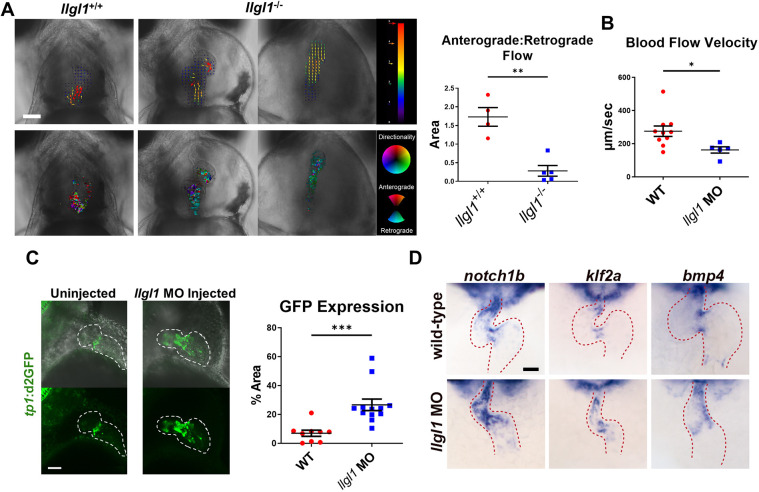
Fig. 5.
Valve morphology and hemodynamics are compromised in llgl1 mutants. (A) Representative images from videos acquired using light microscopy of 99 hpf hearts. A heart from an llgl1−/− embryo with slight pericardial effusion is displayed in the middle, whereas on the right, a heart with severe pericardial effusion is depicted. Above: blood flow was analyzed during diastasis using the particle image velocimetry in Fiji. Arrow vectors show directionality distance moved in pixels. Below: particle image velocimetry (PIV) data were analyzed using the PIV analyzer function in Fiji and qualified for anterograde or retrograde flow. Quantification of anterograde versus retrograde area within the heart chambers during diastasis is depicted in the graph (right). n=4 for llgl1+/+ embryos and n=5 for llgl1−/− embryos. (B) Blood flow velocity measurements analyzed from videos. n=10 for untreated embryos and n=5 for llgl1 morpholino-treated embryos. (C) Representative images from confocal z-stack micrographs of hearts of 2 dpf embryos carrying the tp1 transgenic Notch reporter. The area of cardiac GFP expression is depicted in the graph. n=9 for untreated embryos and n=12 for llgl1 morpholino-treated embryos. (D) Representative images of in situ hybridization of valve markers in 48 hpf fish. Ventral view focusing on the heart (red dashed line). Color model depicts the ventricle in red, atrium in blue, AVC in yellow and venous pole in purple. Two-tailed unpaired Student's t-test. Error bars indicate s.e.m. *P<0.05, **P<0.01, ***P<0.001. Scale bars: 100 µm.