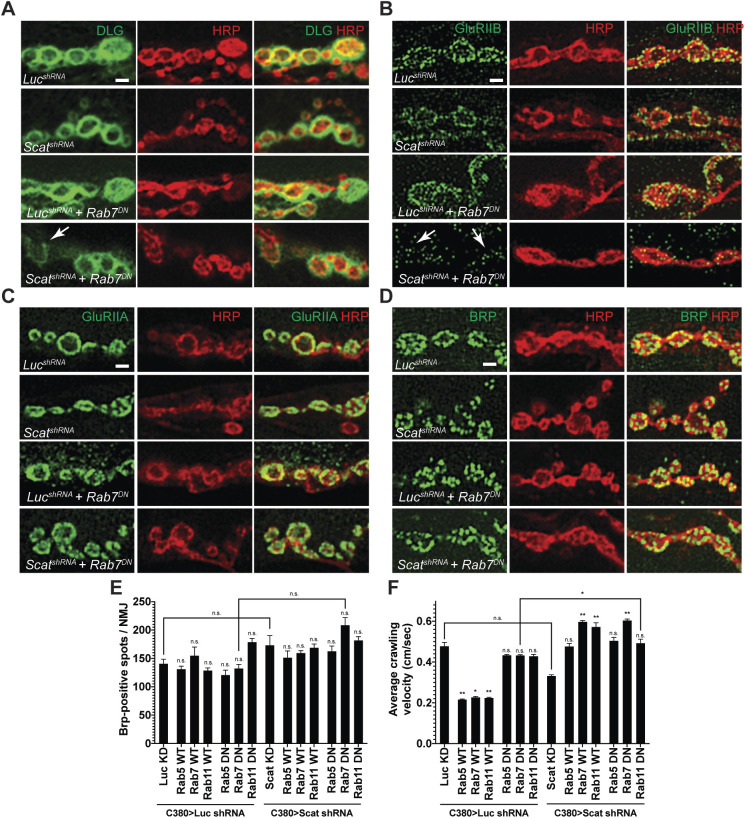
Fig. 6.
scat interacts genetically with rab7 to control the composition of the PSD. Localization of the PSD proteins (A) Dlg, (B) GluRIIB, (C) GluRIIA and the AZ marker (D) Brp to synaptic boutons at muscle 6/7 in body segment A3 in late third instar larvae are shown in green. All NMJs have been counterstained with an antibody targeting HRP (red). Merged images are included to confirm pre- or postsynaptic localization. All images are single focal planes. Synaptic bouton structure has been better preserved here using Bouin's fixative. The arrow in A points to a type 1b bouton with abnormally low levels of Dlg and neighboring boutons have spotty Dlg staining. Arrows in B point to synaptic boutons where GluRIIB localization has been significantly disrupted (compare scatshRNA, Rab7DN to any of the control genotypes). Scale bar: 2.5 µm. (E) Quantification of the number of Brp-positive spots per NMJ. No significant difference was observed in any genotype (N=5 each). Statistical analysis was done using a one-way ANOVA followed by a Holm-Sidak multiple comparison test. (F) Average crawling velocity of third instar larvae for each genotype (N=10 each). MN-specific overexpression of wild-type Rab5, Rab7 and Rab11 alone significantly suppressed larval crawling velocity while overexpression of dominant negative Rabs had no effect. Overexpression of both wild-type and dominant negative forms of all Rabs suppressed the C380>Scat shRNA phenotype. Statistical analysis was done by Kruskal–Wallis followed by a Dunn's multiple comparison test. *P<0.05, **P<0.01.