Abstract
The conspicuous bright golden to orange-reddish coloration of species of the basidiomycete genus Laetiporus is a hallmark feature of their fruiting bodies, known among mushroom hunters as the “chicken of the woods”. This report describes the identification of an eight-domain mono-modular highly reducing polyketide synthase as sole enzyme necessary for laetiporic acid biosynthesis. Heterologous pathway reconstitution in both Aspergillus nidulans and Aspergillus niger verified that LpaA functions as a multi-chain length polyene synthase, which produces a cocktail of laetiporic acids with a methyl-branched C26–C32 main chain. Laetiporic acids show a marked antifungal activity on Aspergillus protoplasts. Given the multiple products of a single biosynthesis enzyme, our work underscores the diversity-oriented character of basidiomycete natural product biosynthesis.
Subject terms: Multienzyme complexes, Fungal physiology
Introduction
Non-terpenoid polyenes are a remarkable class of biologically active basidiomycete natural products. These compounds with up to ten conjugated carbon–carbon double bonds have been attributed to chemical defense: piptoporic acid (Fig. 1), a polyene from Piptoporus australiensis with seven double bonds in conjugation deters fungivorous larvae from feeding on the fruiting bodies [1, 2]. More recently, 18-methyl-19-oxoicosaoctaenoic acid and 20-methyl-21-oxodocosanonaenoic acid (Fig. 1) of a taxonomically undescribed stereaceous basidiomycete, preliminarily referred to as BY1, were shown to inhibit pupation of larvae [3]. Biosynthetically, the respective compounds are polyketides and were instrumental in gaining first insight in polyene biogenesis in Basidiomycota as the BY1 multi-domain highly reducing polyketide synthase (HR-PKS) PPS1 was functionally reconstituted in Aspergillus niger as heterologous host [4].
Fig. 1.
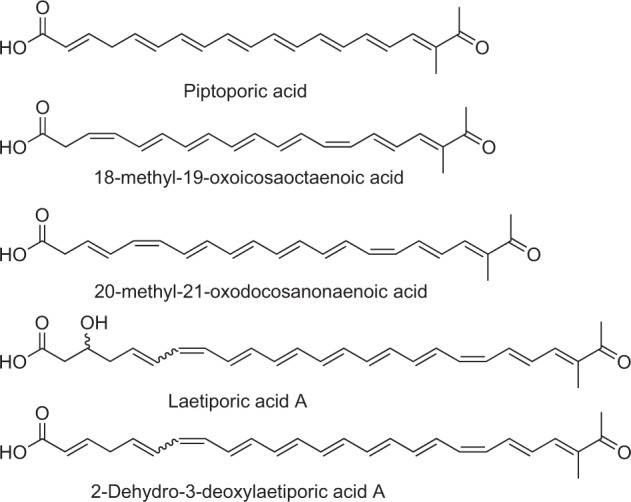
Structures of basidiomycete polyenes. For laetiporic acid A, the predominant isomer is shown (cis-configured double bond C7–C8)
The intense, conspicuous orange color is the signature feature of specimens of the Laetiporus sulphureus species complex, i.e., the “chicken of the woods” fungi. These are brown-rotting bracket mushrooms that have a European and North American distribution and which are commonly found on oak, eucalypt, or willow trees. The coloration is conferred by a blend of polyenes. In previous works, Weber et al. elucidated the structure of laetiporic acid A and its 2 dehydro-3-deoxy derivative (Fig. 1) [5, 6], i.e., two non-terpenoid polyenes that possess a C26 main chain and share the 1-methyl-2-oxo-propylidene moiety with the above basidiomycete polyenes yet show ten conjugated double bonds. The same authors detected even longer putative polyene products, laetiporic acids B and C (with C28 and C30 main chains, respectively) by liquid chromatography and mass spectrometry.
To learn more about the structural diversity of fungal polyenes, including the as yet largely uninvestigated biosynthesis of non-aromatic polyketides in basidiomycetes, we built upon the above previous Laetiporus-related results. Herein, we describe the L. sulphureus enzyme LpaA as a multi-chain length polyene synthase. Heterologous pathway reconstitution in two independent recombinant lpaA-expressing Aspergillus species led to a polyene profile similar to that found in L. sulphureus mycelium and fruiting bodies. We demonstrate that LpaA, i.e., a single HR-PKS, produces a series of compounds with C26–C32 main chain lengths.
Materials and methods
General experimental procedures
Semipreparative HPLC was performed on an Agilent 1260 instrument, equipped with a diode-array detector, UHPLC-MS runs were done on an Agilent 1290 Infinity II chromatograph, interfaced to an Agilent 6130 single quadrupole mass detector. HR-ESI-MS spectra were recorded in positive mode on a Thermo Scientific Exactive Orbitrap instrument. UV/Vis spectra were recorded from λ = 200–700 nm with diode array detectors connected with the respective chromatographs, chromatograms were extracted at λ = 450 nm.
Microbial strains and growth conditions
Escherichia coli XL1-blue was used for routine cloning and was cultivated in LB supplemented with 50 µg ml−1 carbenicillin, if required. Laetiporus sulphureus (s.l.) JMRC SF012599 was provided by the Jena Microbial Resource Collection (JMRC) and was routinely maintained on MEP medium (per liter: malt extract 30 g, soytone peptone 3 g, agar 18 g) for 14 days at 25 °C. To produce laetiporic acids, L. sulphureus was cultivated on 150 YPD agar plates (per liter: yeast extract 10 g, soytone peptone 20 g, d-glucose 20 g, agar 18 g) at 20 °C for 14–21 days. Fruiting bodies of L. sulphureus were collected in Jena, Germany, on willow trees along the Saale river, in September 2019. Aspergillus strains used for transformation were A. niger ATNT16ΔpyrGx24 [7] and A. nidulans FGSC A4.
A. niger transformants tPS01 and tPS02 were cultivated on Aspergillus minimal medium agar plates (AMM) [8] supplemented with 5 mM l-glutamine at 30 °C for 5–7 days. Media for ATNT16ΔpyrGx24 were supplemented with 10 mM uridine. To produce laetiporic acids in recombinant A. niger, transformants tPS01 (vector control) and tPS02 (polyene producer) were pre-cultivated overnight in 30 Erlenmeyer flasks, each filled with 50 ml YPD medium, at 30 °C and 140 rpm. The main culture was a 30 × 1 l fermentation (AMM containing 200 mM d-glucose and 50 mM l-glutamine), inoculated with 50 ml pre-culture each. To induce lpaA expression, 30 mg l−1 doxycycline was added after 18 h, and cultivation was continued for additional 48 h. A. nidulans FGSC A4 and mutant tMG01 were maintained on AMM plates supplemented with 5 mM l-glutamine at 37 °C for 3 days. Plates for tMG01 were supplemented with 0.1 µg ml−1 pyrithiamine hydrobromide. To produce laetiporic acids in A. nidulans, the strains were cultivated in 100 l auto-inducing AMM, prepared with 200 mM ethanol and 10 mM d-glucose as carbon sources, at 30 °C and 140 rpm for 72 h. Details on fungal strains are given in Supplementary Table S1. Aspergillus conidia were harvested with 10 ml sterile water and the suspension was filtered by a cell strainer (40 µm, EASYstrainer). Media were inoculated at a titer of 1 × 106 conidia per milliliter.
cDNA cloning and construction of lpaA expression plasmids
L. sulphureus mycelium was grown in liquid YPD medium at 20 °C and 140 rpm for 7 days, harvested, and ground under liquid nitrogen. RNA was isolated using the SV Total RNA Isolation Kit (Promega). Residual genomic DNA was digested by Baseline-ZERO DNase (Biozym). Reverse transcription was carried out with anchored oligo-dT18 primers and RevertAid Reverse Transcriptase (ThermoFisher). The lpaA coding sequence was PCR-amplified from the first strand reaction, using the oligonucleotides oCL46 and oCL47 (Supplementary Table S2), using method A (Supplementary Table S3). The gel-purified fragment was ligated into pJET1.2 (Thermo) to yield plasmid pCL10 (Supplementary Table S4) which was sequenced (GenBank accession number MT304701) to verify accurate amplification and then served as template for subsequent PCRs. A tag-free version (8190 bp) was expressed in A. nidulans, while in A. niger, a gene for a hexahistidine fusion protein was used (8229 bp).
The lpaA coding sequence was PCR-amplified (method B, Supplementary Table S3) from pCL10 using oMG459 and oMG460 (Supplementary Table S2) in order to introduce PacI sites at either end of the fragment. The A. niger expression vector pSMX2-URA [7], allowing for doxycycline-inducible gene expression, was modified by PCR-mediated ligation (oligonucleotides oMG457/oMG458) to incorporate a PacI restriction site in the multiple cloning site to create vector pPS01 (Supplementary Tables S2, S4). Both the insert and pPS01 were restricted with PacI and ligated to create the lpaA expression vector pPS03. Plasmids pPS01 (vector) and pPS03 (lpaA expression plasmid) were used to transform A. niger.
To construct an alcohol-inducible lpaA expression vector, the vector backbone of plasmid pMD03 [9] as well as the lpaA coding sequence inserted in plasmid pCL10 were amplified (method B, Supplementary Table S3) using oligonucleotides oMG468/oMG469 and oMG471/oMG472, respectively. Both fragments were ligated using the NEBuilder HiFi DNA Assembly Cloning Kit (NEB) to yield lpaA expression plasmid pMG49, which was used to transform A. nidulans. Details of plasmids are described in Supplementary Table S4.
Transformation of Aspergillus species
Protoplast transformation of A. niger and A. nidulans was carried out as previously described [7, 9]. In brief, protoplasts were obtained by incubation of mycelium with VinoflowPro (1.1 g per 20 ml volume) for 4 h in YAT buffer (0.6 M KCl, 50 mM maleic acid, pH 5.5) and 10 µg of plasmid DNA (pPS01 and pPS03 for A. niger, pMG49 for A. nidulans) were used for polyethylene glycol-mediated transformation. A. niger transformants (tPS01 and tPS02) were selected by uracil prototrophy, while A. nidulans transformants (tMG01) were selected by pyrithiamine resistance in presence of 0.1 µg ml−1 pyrithiamine. Integration of the lpaA gene was confirmed by PCR (Supplementary Table S3, methods C and D).
UHPLC analysis of laetiporic acids
Methanolic crude extracts of mycelia from Aspergilli and Laetiporus mycelia and carpophores were centrifuged, filtered, and were subjected to UHPLC measurements. Method B (Supplementary Table S5) was used for initial screening of extracts of positive transformants or selection of fractions containing laetiporic acids during the purification procedure (see below). Method B was also applied for polyene quantification in growth inhibition assays.
Purification of laetiporic acids
One-hundred and fifty Laetiporus agar plates were diced, lyophilized, and extracted with acetone (3 × 5 l). Aspergillus mycelia were collected, washed with water, and lyophilized. Mycelia were ground to a fine powder and extracted six times with methanol and subsequently twice with acetone (50 ml per 1 g dry biomass and extraction).
Aspergillus and Laetiporus extracts were filtered through cellulose round filters and evaporated to dryness. The dry residue was dissolved in 2 l water and repeatedly extracted with a total of 12 l ethyl acetate. The organic phase was evaporated. The residue was dissolved in 400 ml methanol, and 40 ml aliquots were subjected to size exclusion chromatography on Sephadex LH-20 (60 × 4 cm) with methanol as eluent. Three fractions (FI-III) were obtained, containing laetiporic acid (LA)-A, LA-B in F-I, LA-C in F-II, and LA-D and traces of other derivatives in F-III. All fractions were subjected to reversed phase semi-preparative HPLC, using methods C (F-I and F-II) and D (F-III) (Supplementary Table S5). Isolated compounds were lyophilized and dissolved in methanol. Final work up was accomplished under slightly basic conditions (method E for LA-A and LA-B, method F for LA-D, Supplementary Table S5).
To test for photoisomerization, 100 µg ml−1 of Aspergillus-produced laetiporic acids A1, B1, or B2, respectively, or laetiporic acid A1 from L. sulphureus were continuously exposed to light for 24 h (or in the dark for control). After exposure, the solutions were chromatographically analyzed by UHPLC-MS (method A, Supplementary Table S5).
Growth inhibition assays
Cultivations were carried out in triplicate at 30 °C and 140 rpm with a conidial titer of 1 × 106 per ml in 30 ml medium. A. niger tPS01 and tPS02 were cultivated in AMM (+200 mM d-glucose and 50 mM l-glutamine) for 42 h. Doxycycline hydrochloride (0, 7.5, 15, 30, 60, or 120 µg ml−1) was added immediately after inoculation. A. nidulans strains were cultivated in AMM without glucose, but with 200 mM ethanol as sole carbon source (inducing condition) for 72 h. For delayed gene expression, d-glucose (0, 2.5, 5, 10, or 50 mM) was added prior to inoculation. AMM with 200 mM d-glucose (repressing condition) served as negative control. After cultivation, the mycelium was lyophilized, weighted, ground to a fine powder, and extracted with 1 ml methanol for 5 min in an ultrasonic bath. After centrifugation (10 min, 13,000 g), an aliquot of 5 µl was subjected to HPLC analysis (method A, Supplementary Table S5) and polyene signals were manually integrated (λ = 450 nm, tR = 4–8 min). Polyenes were quantified against a calibration curve with respective SEC-purified polyenes as authentic reference standards.
Protoplast toxicity assays
To obtain polyene-containing and control extracts, A. nidulans was cultivated in AMM (+10 mM d-glucose and 200 mM ethanol) at 30 °C and 140 rpm for 72 h. Lyophilized mycelium (5 g) was ground to a powder and extracted five times with 200 ml methanol. Extracts were purified via size exclusion chromatography with Sephadex LH-20 as described above. Fractions were analyzed with UHPLC-MS, and fractions containing laetiporic acids were pooled for tMG01 and added in concentrations from 31 µg ml−1 to 4 mg ml−1. As negative control, appropriate SEC fractions of A. nidulans wild type were pooled accordingly. The eluates were dried under reduced pressure and residues were suspended in sterile YAT buffer.
To produce protoplasts, A. nidulans mycelium was filtered, washed with sterile YAT buffer, and incubated in lysis solution (1.3 g VinoTaste Pro (Novozymes), 0.1 g lysing enzymes from Trichoderma harzianum (Sigma), 0.1 g Yatalase (Takara), in 20 ml YAT buffer) at 30 °C and 70 rpm for 3 h. Protoplasts were filtered through sterile Miracloth, washed with YAT buffer, counted, and diluted to a final titer of 2 × 104 protoplasts per ml. 100 µl (2 × 103 cells) of the cell suspension were gently mixed with the same volume of extracts (A. nidulans FGSCA4 or tMG01), and protoplasts were incubated on ice for 3 h. Suspensions were carefully plated on osmotic AMM plates (with 1.2 M sorbitol, 100 mM d-glucose, 20 mM l-glutamine, pH 6.5) and incubated at room temperature for 96 h. Colony forming units from four independent experiments were counted.
Results
Identification of HR-PKS genes in L. sulphureus
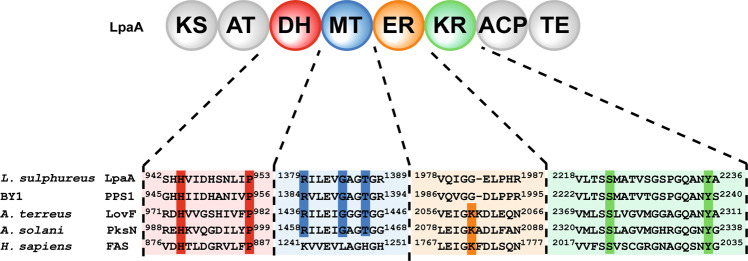
The chemical structures of the laetiporic acids from L. sulphureus and the polyenes isolated from the BY1 mushroom differ in their chain length, yet share a 1-methyl-2-oxo-1-propylidene moiety and a shifted conjugated double bond system, i.e., the double bonds are positioned within the formal acetate units. We therefore used the sequence of PPS1, the only known basidiomycete polyene synthase, as query to browse the published genome of L. sulphureus [10]. We identified two near-identical HR-PKS genes, now collectively referred to as lpaA, that both encode proteins of 2729 amino acids (95% identical and 97% similar amino acids) with a corresponding molecular mass of 296 kDa. The predicted domain architecture ketosynthase–acyltransferase–dehydratase–methyltransferase–enoylreductase–ketoreductase–acyl carrier protein–thioesterase (KS–AT–DH–MT–ER0–KR–ACP–TE, Fig. 2) is consistent with the enzymatic requirements to biosynthesize a methyl branched polyene. The protein LpaA shared 68% identity (81% similarity) to PPS1. Analyses of amino acid sequences identified conserved canonical active sites in all domains, except the ER domain. Consistent with the not fully reduced laetiporic acids, we assumed that the ER domain in LpaA was most likely not functional. Usually, a lysine residue serves as a proton donor during reduction of the enoyl [11]. This role was evident in mammalian fatty acid synthases (K1771) [12], or fully reducing PKSs, such as LovF from Aspergillus terreus (K2060) [13] and Alternaria solani PKSN (K2082) [14]. However, in LpaA, and PPS1 alike, this lysine residue is replaced by glycine (G1982 and G1990), respectively, indicating a structural rather than a catalytic function of the ER (Fig. 2).
Fig. 2.
Domain setup of LpaA. Domain abbreviations: β-keto synthase (KS), acyltransferase (AT), dehydratase (DH), methyltransferase (MT), enoyl reductase (ER), β-keto reductase (KR), acyl carrier protein (ACP), thioesterase (TE). Below: amino acid sequence alignments of the active site of the respective domains. Conserved residues are highlighted with vertical color-coded bars. BY1 mushroom polyene synthase PPS1 (NCBI accession #: KX819293.1); A. terreus LovF: Aspergillus terreus lovastatin diketide synthase (Q9Y7D5.1); A. solani PksN: Alternaria solani alternapyrone synthase (BAD83684.1); H. sapiens FAS: human fatty acid synthase (NP_004095.4). A MT domain is not present in the human FAS. The ER domain is inactive in LpaA and PPS1. A TE domain is not present in LovF and PksN
Construction of recombinant, lpaA-expressing Aspergillus niger
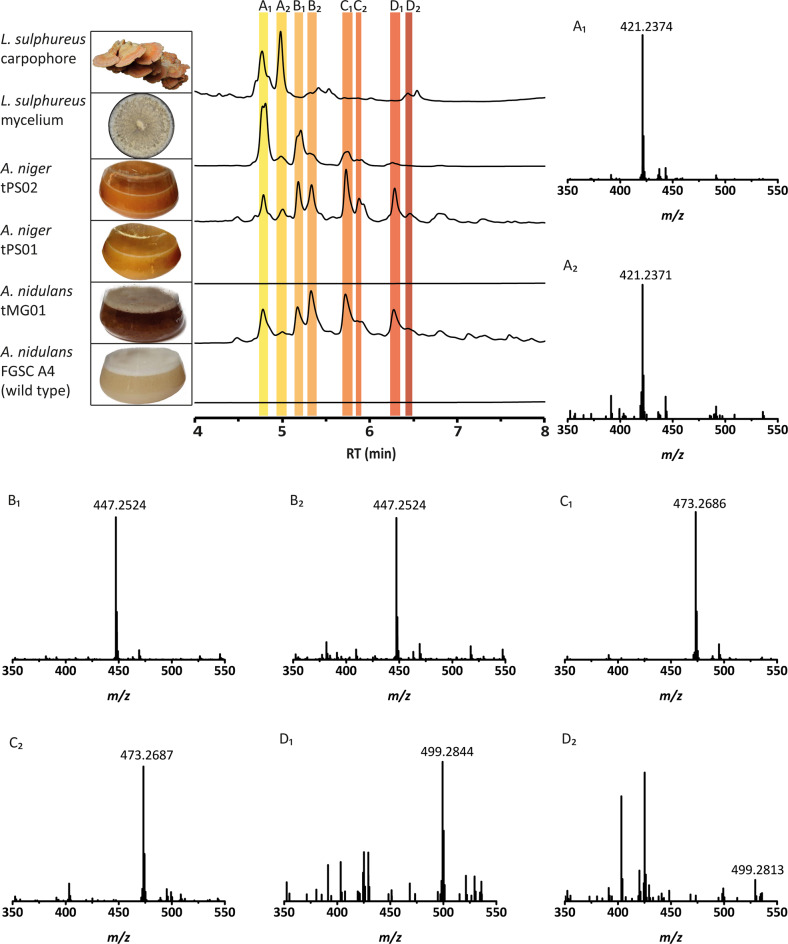
L. sulphureus produces laetiporic acid A in fruiting bodies as well as in mycelia, as described [5] and confirmed in our study by UHPLC-MS and HR-MS (Fig. 3). To investigate the function of lpaA, the full-length gene was amplified from cDNA of mycelium, sequenced, and ligated to plasmid pSMX2-URA to allow for a tunable, doxycycline-dependent expression in the host A. niger ATNT16ΔpyrGx24 [7]. The fungus was transformed with a lpaA-expressing plasmid (pPS03) as well as an empty vector (pPS01) as control, yielding A. niger tPS02 (lpaA-expressing) and control strain A. niger tPS01 (Supplementary Table S1) The full-length integration of the constructs into the genome was verified by PCR (Supplementary Fig. S1). Both tPS01 and tPS02, were initially cultivated in presence and absence of doxycycline, but immediate induction led to poor growth of tPS02, while it did not impact growth of tPS01. To produce sufficient biomass for further chromatographic analysis, the cultures were induced with doxycycline (30 mg l−1) 18 h post inoculation. The cultivation was continued for another 48 h during which mycelium of tPS02 turned orange (Fig. 3), while both the culture supernatant and the control strain tPS01 did not change their color.
Fig. 3.
UHPLC profiles of metabolic extracts from cultures of L. sulphureus, lpaA-expressing Aspergillus strains and their respective control strains. Representative pictures of the fruiting bodies or cultures are presented (left). UHPLC profiles (top to bottom) of methanolic extracts from L. sulphureus carpophores, vegetative mycelium of L. sulphureus, A. niger tPS02 (lpaA-expressing), A. niger tPS01 (vector control), A. nidulans tMG01 (lpaA-expressing) and A. nidulans FGSC4 wildtype strain. Peaks for laetiporic acids A1–D2 are indicated by colored bars. Chromatograms were extracted at λ = 450 nm. Minor UV-absorbing compounds surrounding laetiporic acids A1–D2 are present that have not been identified, but were absent in the controls. Respective high-resolution mass spectra are given for laetiporic acids A1–D2 purified from A. niger. Polyene peak intensities of mass spectra range between 6 × 105 (LA-D2) and 1.8 × 107 (LA-B1). Please refer to Table 1 for MS² data. LA-D2 was produced in insufficient quantities for further analysis
Identification of laetiporic acids from lpaA-expressing Aspergillus strains
Mycelia of the induced transformants tPS01 and tPS02 were extracted with methanol. A subsequent UHPLC-MS analysis revealed various novel signals in tPS02 at λ = 450 nm, compared to the control tPS01. UV/Vis spectra of the new peaks showed absorption maxima between λ = 430–480 nm (Supplementary Fig. S2) and were reminiscent of the spectra of BY1 polyenes 18-methyl-19-oxoicosaoctaenoic acid and 20-methyl-21-oxodocosanonaenoic acid [4]. We therefore hypothesized the signals may reflect the presence of polyenes due to activity of LpaA in vivo. Curiously, a series of masses was found which followed a regular pattern in increments of m/z + 26, i.e., C2H2 (Table 1). The eight most abundant peaks (Fig. 3) were purified by size exclusion and semipreparative liquid chromatography and were finally subjected to HR-MS/MS detecting over a range between m/z 200 and 1400. Three detected signals are compatible with the masses of laetiporic acids A1 (m/z 421 [M + H]+), B1 (m/z 447 [M + H]+) and C1 (m/z 473 [M + H]+) as previously described [5]. Furthermore, we found a compound with m/z 499 [M + H]+ which we preliminarily refer to as laetiporic acid D1. The masses point to main chain lengths of C26–C32. Surprisingly, this set of masses appeared duplicated (laetiporic acids A2, (m/z 421 [M + H]+), B2 (m/z 447 [M + H]+), C2 (m/z 473 [M + H]+) and D2 (m/z 499 [M + H]+) which may reflect a cis/trans-isomerization of the polyene backbone. A 7-cis isomer corresponding to the 7-trans laetiporic acid A has previously been isolated from L. sulphureus [5, 6]. Additional confirmation that laetiporic acids had been produced in the transgenic Aspergilli came from comparative LC-MS2 analysis of laetiporic acid A1 with a standard isolated from mycelium of the original producer L. sulphureus (Figures S3 and S4) which yielded virtually identical signal patterns. Laetiporic acids were described as stable compounds [5]. To confirm these prior results, we exposed laetiporic acids A1, (isolated both from Aspergillus niger tPS02 and L. sulphureus), B1, and B2, respectively, to light for 24 h, or kept them in the dark for control. Chromatographic analysis did not indicate new signals, which supports the notion of laetiporic acids as stable compounds (Supplementary Fig. S5). In return, this finding suggests that the polyene diversity is an inherent feature of Laetiporus polyketide biosynthesis.
Table 1.
MS and MS2 data of laetiporic acid pairs A1/A2–D1/D2
| Compound | tR (min) | Formula | Neutral mass [M] | Found parental ion [M + H]+ | MS2 specific ions | ||
|---|---|---|---|---|---|---|---|
| Ion mass | Formula | Origin | |||||
| LA-A1/A2 | 4.7/4.9 | C27H32O4 | 420.2301 | 421.2374/421.2371 | 403.227 | C27H31O3 | [M-H2O + H]+ |
| 385.216 | C27H29O2 | [M-2 H2O + H]+ | |||||
| 359.236 | C26H31O | [M-H2O-CO2 + H]+ | |||||
| 343.190 | C21H27O4 | [M-C6H6 + H]+ | |||||
| 245.132 | C19H17 | [M-C8H8-H8O4 + H]+ | |||||
| 219.117 | C17H15 | [M-C10H10-H8O4 + H]+ | |||||
| 193.101 | C15H13 | [M-C12H12-H8O4 + H]+ | |||||
| 131.086 | C10H11 | [C10H10 + H]+ | |||||
| 109.065 | C7H10O | [C7H9O + H]+ | |||||
| 105.070 | C8H9 | [C8H8 + H]+ | |||||
| 79.054 | C6H7 | [C6H6 + H]+ | |||||
| LA-B1/B2 | 5.1/5.3 | C29H34O4 | 446.2457 | 447.2524/447.2524 | 429.242 | C29H33O3 | [M-H2O + H]+ |
| 411.231 | C29H31O2 | [M-2 H2O + H]+ | |||||
| 369.204 | C23H29O4 | [M-C6H6 + H]+ | |||||
| 351.195 | C23H27O3 | [M-C6H6-H2O + H]+ | |||||
| 312.172 | C20H24O3 | [M-C9H9-H2O + H]+ | |||||
| 271.148 | C21H19 | [M-C8H8-H8O4 + H]+ | |||||
| 245.132 | C19H17 | [M-C10H10-H8O4 + H]+ | |||||
| 219.117 | C17H15 | [M-C12H12-H8O4 + H]+ | |||||
| 193.101 | C15H13 | [M-C14H14-H8O4 + H]+ | |||||
| 131.086 | C10H11 | [C10H10 + H]+ | |||||
| 109.065 | C7H10O | [C7H9O + H]+ | |||||
| 105.070 | C8H9 | [C8H8 + H]+ | |||||
| 79.054 | C6H7 | [C6H6 + H]+ | |||||
| LA-C1/C2 | 5.7/5.9 | C31H36O4 | 472.2614 | 473.2686/473.2687 | 455.258 | C31H35O3 | [M-H2O + H]+ |
| 395.219 | C25H31O4 | [M-C6H6 + H]+ | |||||
| 338.188 | C22H26O3 | [M-C9H9-H2O + H]+ | |||||
| 245.132 | C19H17 | [M-C12H12-H8O4 + H]+ | |||||
| 219.117 | C17H15 | [M-C14H14-H8O4 + H]+ | |||||
| 193.101 | C15H13 | [M-C16H16 -H8O4 + H]+ | |||||
| 131.086 | C10H11 | [C10H10 + H]+ | |||||
| 109.065 | C7H10O | [C7H9O + H]+ | |||||
| 105.070 | C8H9 | [C8H8 + H]+ | |||||
| 79.054 | C6H7 | [C6H6 + H]+ | |||||
| LA-D1 | 6.2 | C33H38O4 | 498.2770 | 499.2844 | 481.2742 | C33H37O3 | [M-H2O + H]+ |
| 421.2373 | C27H33O4 | [M-C6H6 + H]+ | |||||
| 364.203 | C24H28O3 | [M-C9H9-H2O + H]+ | |||||
| 297.163 | C23H21 | [M-C10H10-H8O4 + H]+ | |||||
| 219.117 | C17H15 | [M-C16H16-H8O4 + H]+ | |||||
| 193.101 | C15H13 | [M-C18H18-H8O4 + H]+ | |||||
| 131.086 | C10H11 | [C10H10 + H]+ | |||||
| 109.065 | C7H10O | [C7H9O + H]+ | |||||
| 105.070 | C8H9 | [C8H8 + H]+ | |||||
| 79.054 | C6H7 | [C6H6 + H]+ | |||||
| LA-D2 | 6.4 | C33H38O4 | 498.2770 | 499.2832 | not determined. | ||
The notorious very poor solubility of laetiporic acids (<1 mg ml−1) in organic solvents [5] such as methanol, chloroform, dichloromethane, cyclohexane, butanol, or acetone, combined with the structural similarity of the various compounds prevented recording of unambiguous NMR spectra to confirm structures and, in particular, double bond positions and configurations. For further characterization, we therefore resorted to MS2 experiments (Table 1, Supplementary Fig. S3) for seven out of eight metabolites. As expected for β-hydroxy acids, MS2 spectra revealed a consistent prevalence for dehydration (m/z [M−18 + H]+) and decarboxylation (m/z [M−44 + H]+) during fragmentation of all identified laetiporic acids. Moreover, the elimination of aromatic rings such as benzene (m/z 79 [M + H]+) by electrocyclic butyl ring contraction provided a defined MS fingerprint for the linear polyene structure of laetiporic acids resulting in MS2 fragments of m/z [M−78 + H]+. This is in agreement with electrocyclic aromatic elimination of toluene and xylene observed in ESI-based MS2 fragmentation of polyene-like carotenoids [15, 16].
Beyond these eight laetiporic acids, we detected traces of another set of very minor signals that overlapped with the major peaks. The molecular masses of the latter indicated dehydrated analogs (m/z [M−18 + H]+) of laetiporic acids A–D to give presumably 2-dehydro-3-deoxylaetiporic acids A–D (m/z 403, 429, 455, and 481 [M + H]+, respectively). These compounds were produced in amounts that were insufficient for further analysis. Still, a very similar phenomenon was recognized previously when 2-dehydro-3-deoxylaetiporic acid A (Fig. 1) was identified as a side product from L. sulphureus [6].
The above findings are remarkable in that one single PKS makes polyenes of (i) different chain length (C26–C32) and (ii) different degree of hydroxylation. Although precedence exists for variable chain lengths of HR-PKS PPS1 (C20–C22) of the basidiomycete BY1 [4] and T-toxin (C35–C45) the high virulence determinant of the ascomycetous maize pathogen Cochliobolus heterostrophus [17], the wide spectrum of metabolites produced by LpaA in A. niger might be a host-specific effect. Laetiporic acids A1 and A2 are the major metabolites isolated from the fruiting body or the mycelium of L. sulphureus [6]. In contrast, heterologous production predominantly resulted in laetiporic acids B and C (Fig. 3). Therefore, A. nidulans FGSC A4 was tested as alternative host and transformed with plasmid pMG49 in which the lpaA gene is controlled by the ethanol-inducible, but glucose-repressible alcohol-dehydrogenase promoter (PalcA, Supplementary Tables S1, S4 and Supplementary Fig. S1) [18]. The resulting transformant tMG01 and the wild type show comparable growth in presence of d-glucose. However, growth was impeded when ethanol was used as sole carbon source indicating similar toxic effects of laetiporic acids as observed for A. niger. When supplemented with 10 mM d-glucose, the mutant showed a strongly retarded growth and its mycelium turned orange to intense red (Fig. 3). Mycelial extracts from tMG01 were analyzed by UHPLC-MS and revealed a similar chromatographic profile as observed for the lpaA-expressing A. niger mutant tPS02. In contrast, no signals at λ = 450 nm were detectable in the parental strain. In summary, these results confirmed laetiporic acids A1–D2 as products of LpaA and point to an antifungal activity of laetiporic acids when produced in situ.
Antifungal activity of laetiporic acids
To follow up on the impeded growth of polyene-producing Aspergilli, we assessed the antifungal activity of the produced cocktail. The agar diffusion assay—a widely used technique in antimicrobial susceptibility testing—relies on water soluble and diffusible substances, which cannot be assumed for laetiporic acids. As an alternative dual strategy, we tested the bioactivity of polyenes (i) in situ during production in A. niger and A. nidulans (Fig. 4), and (ii) directly on A. nidulans protoplasts (Fig. 5). A. niger strains tPS01 (control) and tPS02 (lpaA-expressing) were cultivated in the absence or presence of 7.5–120 µg ml−1 doxycycline to induce gene expression dose-dependently [19]. While doxycycline hardly inhibited growth at 30–120 µg ml−1 in the tPS01 control, tPS02 was strongly impaired, and had produced less biomass (Fig. 4). Moreover, growth inhibition correlated with the dose-dependent intrinsic polyene concentration which reached its maximum at 7.5 µg per mg biomass at 120 µg ml−1 doxycycline. In a complementary experiment, the lpaA-expressing strain A. nidulans tMG01 and its parental strain were cultivated in AMM with 200 mM ethanol as carbon source, and variable amounts of glucose (2.5–50 mM) were added prior to inoculation to dose-dependently repress gene expression. Growth of the parental strain was not affected. In contrast, but similarly to what was observed for A. niger, growth inhibition was strongest when the highest intrinsic polyene concentration was reached (110 µg mg−1 polyenes at 2.5 mM d-glucose). Moreover, without repressing d-glucose (that is, 200 mM ethanol as sole carbon source), spores from tMG01 were arrested in the state of germination, indicating a strong antifungal activity. In a parallel experiment, the LpaA-produced cocktail of multi-chain length laetiporic acids was added to A. nidulans wild type protoplasts. Based on the number of colony-forming units, we observed a significantly reduced protoplast viability with undiluted and 1:5 diluted extract, compared to A. nidulans wild type extracts (Fig. 5).
Fig. 4.
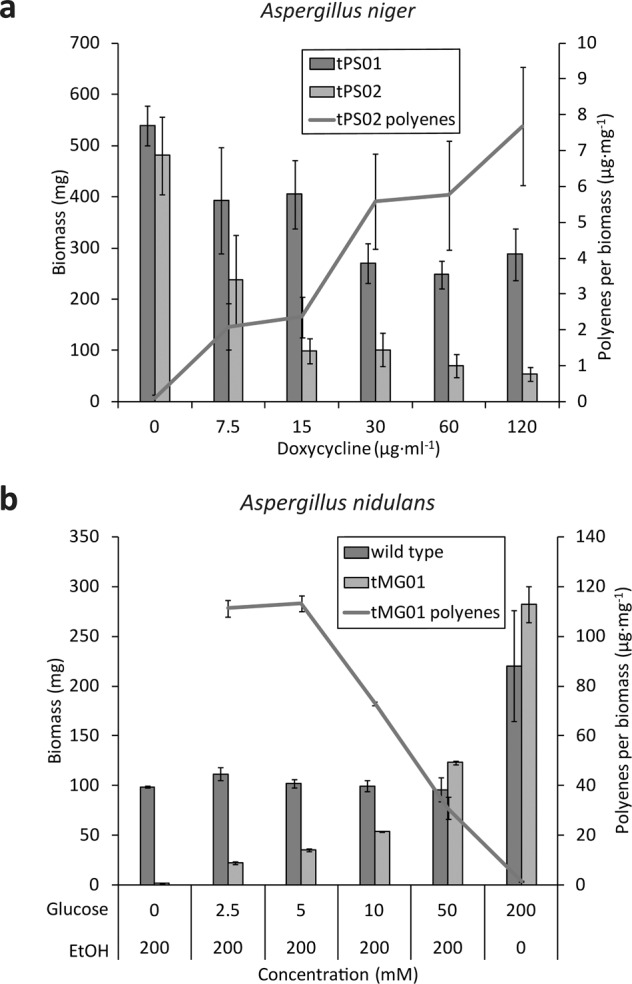
Antifungal activity of laetiporic acids produced in situ by Aspergillus niger and Aspergillus nidulans. Fungal dry biomass (in mg) was determined for A. niger tPS02 (lpaA-expressing) and tPS01 (vector control) (a) or A. nidulans tMG01 (lpaA-expressing) and FGSC A4 wildtype strain (b), respectively. Polyene production was induced by adding doxycycline (7.5–120 µg ml−1) in A. niger and repressed by d-glucose (2.5–50 mM) in A. nidulans. Cultures without inducer, i.e., doxycycline for A. niger or ethanol in A. nidulans, served as negative controls. Intracellular polyene concentration is given in µg per mg dry fungal biomass. Polyene production is increased 15-fold in A. nidulans, compared to A. niger, resulting in a more intense coloration and an antifungal effect (no growth under non-repressing conditions). Hence, polyene content could not be determined under this condition. Bars indicate the standard deviation (n = 3)
Fig. 5.
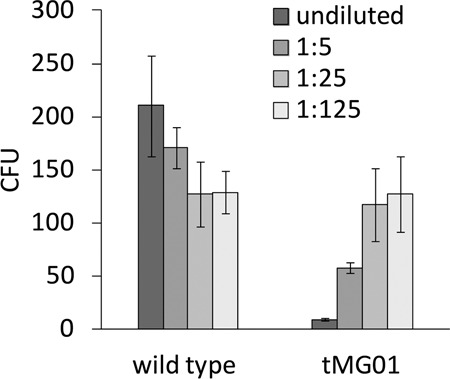
Antifungal activity on Aspergillus nidulans protoplasts. Extracts of A. nidulans wild type (negative control) and tMG01 were pre-purified, and added undiluted or diluted 1:5, 1:25, and 1:125, respectively, to a protoplast suspension. The undiluted tMG01 extract contained polyenes at a concentration of 4 mg ml−1. Bars indicate the standard deviation (n = 4); CFU: colony forming units
Discussion
Conjugated double bonds are a widespread feature of biologically active natural products, among them the leukotrienes, carotenoids, and clinically used antimicrobially active polyenes, such as nystatin and amphotericin. In the context of basidiomycetes, polyenes are remarkable in that they occur erratically and in taxonomically unrelated groups, among them the Russulales [4] and the Polyporales [5, 6]. Knowledge on the biosynthesis of basidiomycete highly reduced polyketide is scanty. To date, PPS1, the characterized polyene synthase of BY1, a stereaceous basidiomycete [4], is the only reported example. To exclude host-specific effects, we relied on two independent heterologous systems: in one system gene expression is mediated by increasing concentrations of an inductor (doxycycline), while in the second system, the ethanol-mediated induction was attenuable by increasing d-glucose concentrations as repressor. Yet, the polyene profiles were highly congruent, independent of the used strategy.
The combined results from PPS1 and the Laetiporus enzyme LpaA reveal common features between these two synthases from unrelated species. Remarkably, polyenes of variable chain length seem to be an intrinsic property of either synthase, yet more pronounced with LpaA, as chain lengths varied between C26 and C32, while PPS1 produces polyenes with C20–C22 main chains. Further, double bond shift (i.e., positioned within formal acetate units, rather than between them) is a shared intrinsic property of LpaA and PPS1. In the case of 2-dehydro-3-deoxylaetiporic acid A (Fig. 1), one double bond remains at the canonical α,β-position, although this may be a secondary effect as well and the consequence of water elimination. Finally, either enzyme showed a preference for a particular chain length, in the case of LpaA primarily C28 and C30 main chains (i.e., laetiporic acids B and C).
Remarkably, LpaA, i.e., a single enzyme, produces an entire set of compounds, which once again supports both the notion of a diversity-oriented secondary metabolism [20] and an emerging concept of multi-product PKSs. Described examples include, e.g., PKS1 of Colletotrichum lagenarium that produces tetra- to hexaketides [21] and the A. terreus polyketide synthase TerA, which makes tri- to pentaketides [22]. Building upon prior results by Weber et al., we here show that a single enzyme produces four chains, which provides the first layer of diversification and underscores that LpaA has been evolved for diversity, rather than specificity. The second layer to generate diversity is elimination of a formal water molecule from positions C-3 and C-4. Elimination of water is a natural, previously described process [5, 6], which is confirmed by our results. A third dimension to generate diversity is the configuration of double bonds.
Knowledge about the ecological role of basidiomycete polyenes is incomplete, yet inhibition of insect larvae was shown by previous works [1, 2, 4]. Our results on both Aspergillus protoplasts and the transgenic Aspergilli add antifungal activity to the biological effects of basidiomycete polyenes. Given the lipophilic character and the length of Laetiporus polyenes, activity may be exerted through interference with membranes. However, the assay system included wall-less cells and a production in situ. Therefore, future research is warranted to determine, if intact cell walls protect mycelium from being inhibited. Still, our results confirm the Basidiomycota as prolific producers of antibiotic compounds [23]. Hallmark examples are the agriculturally used fungicides derived from strobilurine [24], or the clinically used derivatives of the antibacterial pleuromutilin [25]. The results presented here contribute to efforts, reflected by this special issue of the Journal of Antibiotics, to understand this aspect of basidiomycete biology more profoundly.
Supplementary information
Acknowledgements
We thank Andrea Perner (Hans-Knöll-Institute Jena) for recording high-resolution mass spectra. We thank Patricia Hübel and Erik Fischer (Friedrich-Schiller-University Jena) for additional compound purification. We thank Matthias Brock, University of Nottingham for providing A. niger ATNT16ΔpyrGx24 and plasmid pSMX2-URA. CL acknowledges a doctoral fellowship by the International Leibniz Research School (ILRS) for Microbial Interactions. DH is supported by the DFG Collaborative Research Center ChemBioSys 1127.
Funding
Open access funding provided by Projekt DEAL.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41429-020-00362-6) contains supplementary material, which is available to authorized users.
References
- 1.Gill M. Polyolefinic 18-methyl-19-oxoicosenoic acid pigments from the fungus Piptoporus australiensis (Wakefield) Cunningham. J Chem Soc Perkin Trans. 1982;1:1449–53. doi: 10.1039/p19820001449. [DOI] [Google Scholar]
- 2.Gill M, Steglich W. Pigments of fungi (Macromycetes). In: Herz W, Grisebach H, Kirby GW, Tamm C. editors. Progress in the chemistry of organic natural products. Vienna, New York: Springer; 1987. [DOI] [PubMed]
- 3.Schwenk D, Nett M, Dahse HM, Horn U, Blanchette RA, Hoffmeister D. Injury-induced biosynthesis of methyl-branched polyene pigments in a white-rotting basidiomycete. J Nat Prod. 2014;77:2658–63. doi: 10.1021/np500552a. [DOI] [PubMed] [Google Scholar]
- 4.Brandt P, García-Altares M, Nett M, Hertweck C, Hoffmeister D. Induced chemical defense of a mushroom by a double-bond-shifting polyene synthase. Angew Chem Int Ed. 2017;56:5937–41.. doi: 10.1002/anie.201700767. [DOI] [PubMed] [Google Scholar]
- 5.Weber RWS, Mucci A, Davoli P. Laetiporic acid, a new polyene pigment from the wood-rotting basidiomycete Laetiporus sulphureus (Polyporales, Fungi) Tetrahedron Lett. 2004;45:1075–8. doi: 10.1016/j.tetlet.2003.11.073. [DOI] [Google Scholar]
- 6.Davoli P, Mucci A, Schenetti L, Weber RW. Laetiporic acids, a family of non-carotenoid polyene pigments from fruit-bodies and liquid cultures of Laetiporus sulphureus (Polyporales, Fungi) Phytochemistry. 2005;66:817–23. doi: 10.1016/j.phytochem.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 7.Geib E, Baldeweg F, Doerfer M, Nett M, Brock M. Cross-chemistry leads to product diversity from atromentin synthetases in Aspergilli from section Nigri. Cell Chem Biol. 2019;26:223–34.. doi: 10.1016/j.chembiol.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 8.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AWJ. The genetics of Aspergillus nidulans. In: Demerec M. Advances in genetics. 5th ed. New York, N.Y.: Academic Press; 1953. p. 141–238. [DOI] [PubMed]
- 9.Dörfer M, et al. Melleolides impact fungal translation via elongation factor 2. Org Biomol Chem. 2019;17:4906–16.. doi: 10.1039/C9OB00562E. [DOI] [PubMed] [Google Scholar]
- 10.Nagy LG, et al. Comparative genomics of early-diverging mushroom-forming fungi provides insights into the origins of lignocellulose decay capabilities. Mol Biol Evol. 2016;33:959–70. doi: 10.1093/molbev/msv337. [DOI] [PubMed] [Google Scholar]
- 11.Kwan DH, Leadlay PF. Mutagenesis of a modular polyketide synthase enoylreductase domain reveals insights into catalysis and stereospecificity. ACS Chem Biol. 2010;5:829–38.. doi: 10.1021/cb100175a. [DOI] [PubMed] [Google Scholar]
- 12.Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–22. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy J, et al. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284:1368–72. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 14.Fujii I, Yoshida N, Shimomaki S, Oikawa H, Ebizuka Y. An iterative type I polyketide synthase PKSN catalyzes synthesis of the decaketide alternapyrone with regio-specific octa-methylation. Chem Biol. 2005;12:1301–09. doi: 10.1016/j.chembiol.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 15.Neto FC, et al. Re‐investigation of the fragmentation of protonated carotenoids by electrospray ionization and nanospray tandem mass spectrometry. Rapid Commun Mass Spectrom. 2016;30:1540–8. doi: 10.1002/rcm.7589. [DOI] [PubMed] [Google Scholar]
- 16.Rivera SM, Christou P, Canela-Garayoa R. Identification of carotenoids using mass spectrometry. Mass Spectrom Rev. 2014;33:353–72. doi: 10.1002/mas.21390. [DOI] [PubMed] [Google Scholar]
- 17.Baker SE, et al. Two polyketide synthase-encoding genes are required for biosynthesis of the polyketide virulence factor, T-toxin, by Cochliobolus heterostrophus. Mol Plant Microbe Int. 2006;19:139–49. doi: 10.1094/MPMI-19-0139. [DOI] [PubMed] [Google Scholar]
- 18.Kulmburg P, Mathieu M, Dowzer C, Kelly J, Felenbok B. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol Microbiol. 1993;7:847–57.. doi: 10.1111/j.1365-2958.1993.tb01175.x. [DOI] [PubMed] [Google Scholar]
- 19.Geib E, Brock M. ATNT: an enhanced system for expression of polycistronic secondary metabolite gene clusters in Aspergillus niger. Fungal Biol Biotechnol. 2017;4:13. doi: 10.1186/s40694-017-0042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firn RD, Jones CG. Natural products—a simple model to explain chemical diversity. Nat Prod Rep. 2003;20:382–91. doi: 10.1039/b208815k. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe A, Ebizuka Y. Unprecedented mechanism of chain length determination in fungal aromatic polyketide synthases. Chem Biol. 2004;11:1101–6. doi: 10.1016/j.chembiol.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 22.Zaehle C, et al. Terrein biosynthesis in Aspergillus terreus and its impact on phytotoxicity. Chem Biol. 2014;21:719–31. doi: 10.1016/j.chembiol.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Schüffler A, Anke T. Fungal natural products in research and development. Nat Prod Rep. 2014;31:1425–48.. doi: 10.1039/C4NP00060A. [DOI] [PubMed] [Google Scholar]
- 24.Sauter H, Steglich W, Anke T. Strobilurins: evolution of a new class of active substances. Angew Chem Int Ed. 1999;38:1328–49. doi: 10.1002/(SICI)1521-3773(19990517)38:10<1328::AID-ANIE1328>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 25.Novak R, Shlaes DM. The pleuromutilin antibiotics: a new class for human use. Curr Opin Investig Drugs. 2010;11:182–91. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.