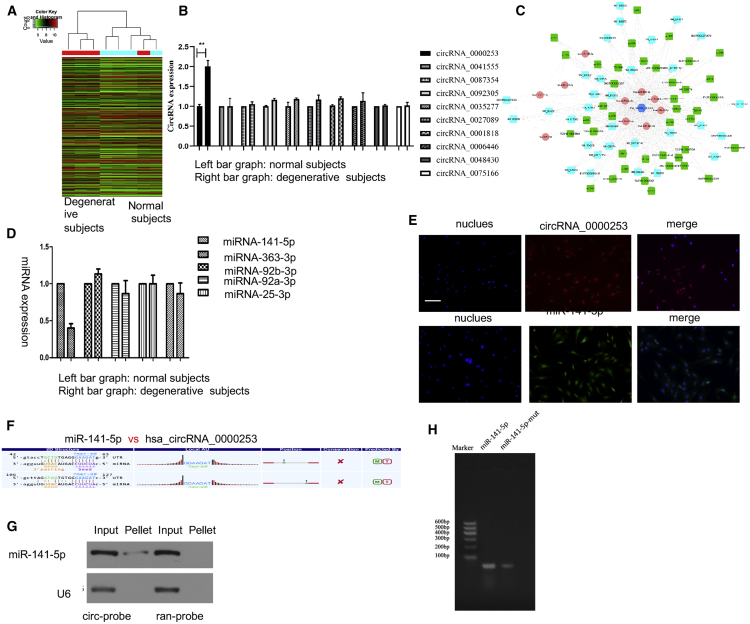
Figure 3.
circRNA_0000253 Competitively Adsorbs microRNA (miRNA)-141-5p
(A) The circRNAs differentially expressed in the nucleus pulposus (NP) tissue. (B) The top 10 upregulated circRNAs were verified by qPCR in degenerative and normal NP exosomes. (C) The possible sponged miRNAs of circRNA_0000253. (D) The possible sponged miRNAs were verified by qPCR in degenerative and normal NP exosomes. (E) RNA fluorescence in situ hybridization (FISH) was used to detect the colocalization of circRNA_0000253 and miRNA-141-5p in NPCs. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 100 μm. (F) The binding sequence diagram of circRNA_0000253 and miRNA-141-5p. (G) miRNA-141-5p was pulled down by the circular probe (circ-probe) but not the random probe (ran-probe), and the levels of miRNA-141-5p were detected by northern blot. Input, 20% samples were loaded; pellet, all samples were loaded. (H) Pull-down assay analysis confirmed a greater enrichment of circRNA_0000253 in the miRNA-141-5p-captured fraction compared with the introduction of the miRNA-141-5p mutation that disrupted the binding site of miRNA-141-5p in circRNA_0000253.