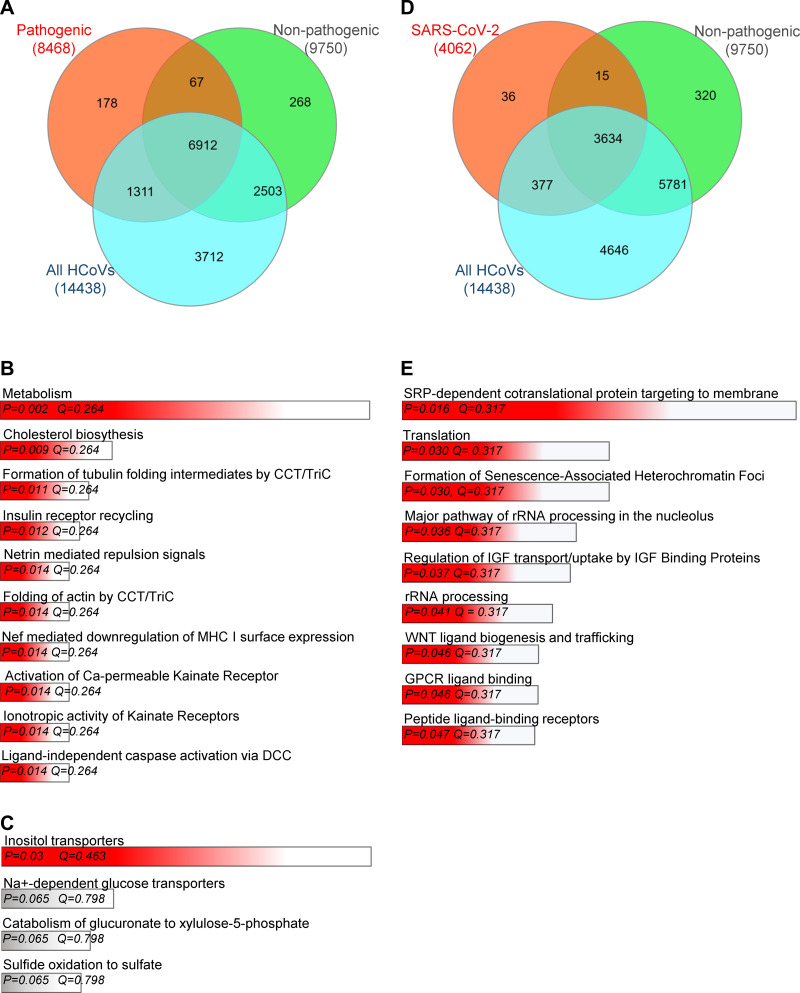
Fig. 2.
The impact of the pathogenic human coronaviruses (HCoV)-dependent modulation of hosts miRNA profiles and their potential consequences on cellular mRNA levels. A: Venn diagram (43) represents the potential general distribution of human mRNAs that could be affected by the miRNAs that target the pathogenic (orange), the nonpathogenic (green) HCoVs, and all seven coronaviruses (blue). The hypothesis is that the miRNA sponge effect used by the individual viruses could lead to an increase in these human mRNAs since these are also targets of these miRNAs. B: the Gene Ontology assignment of the cellular functions of mRNAs defines very specific targets for the miRNAs potentially regulated by the pathogenic HCoVs as assigned by the Enrichr web server (21, 57). C: the Gene Ontology assignment of the cellular function of mRNAs defines specific targets for the miRNAs potentially regulated by the harmless HCoVs. D: the Venn diagram represents the general distribution of mRNAs that are targets of miRNAs modulated by severe acute respiratory coronavirus 2 (SARS-CoV-2) (orange) and the nonpathogenic (green) HCoVs, and all 7 HCoVs (blue). E: the Gene Ontology assignment of the cellular functions of mRNAs that are high probability targets for the miRNAs potentially modulated by the SARS-CoV-2, as assigned by the Enrichr web server (21, 57). All of the mRNA targets were predicted with the miRDIP database (99) with only the top 1% of the most probable targets considered. The predicted targets lists are provided in Supplemental Data Set S2 (https://doi.org/10.5281/zenodo.3966446). The red bar color depicts the P value less than 0.05. The longer bars have the lower P values. The false discovery rate is provided as a Q value.