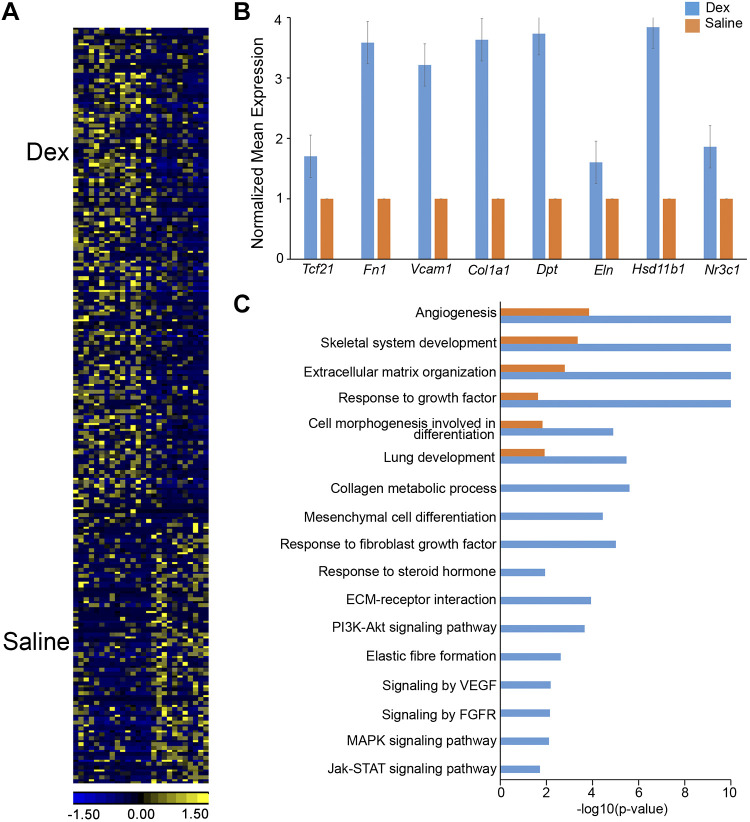
Fig. 5.
Dexamethasone induced gene expression and associated bioprocesses in lung matrix fibroblasts. Dams were injected with 2 mg/mL dexamethasone (Dex) or saline in vivo 24 h before euthanize at embryonic day (E)18.5. Single cell RNA was prepared from lungs of embryos (n = 4–6 pooled lungs per condition). Cell type mapping and signature gene identification were performed using SINCERA pipeline (27). Matrix fibroblasts (MFBs) and associated signature genes were used for heatmap and functional enrichment analysis. A: heatmap of differentially expressed genes (P value < 0.01 and fold change ≥ 1.5) in lung MFBs treated with Dex (n = 15 cells) versus saline (n = 11 cells) at E18.5. B: dexamethasone increased expression of MFB signature genes (blue bars) compared with saline (orange bars). Mean expression was normalized to saline control for each gene. All genes passed Student’s t test (P < 0.05, fold change>1.5) C: top enriched bioprocesses and pathways in genes increased by Dex (blue bars) compared with Saline (orange bars) in MFB at E18.5. Hypergeometric test was used for gene set enrichment analysis with significance at P < 0.001. PI3K, phosphatidylinositol 3-kinase; ECM, extracellular matrix.