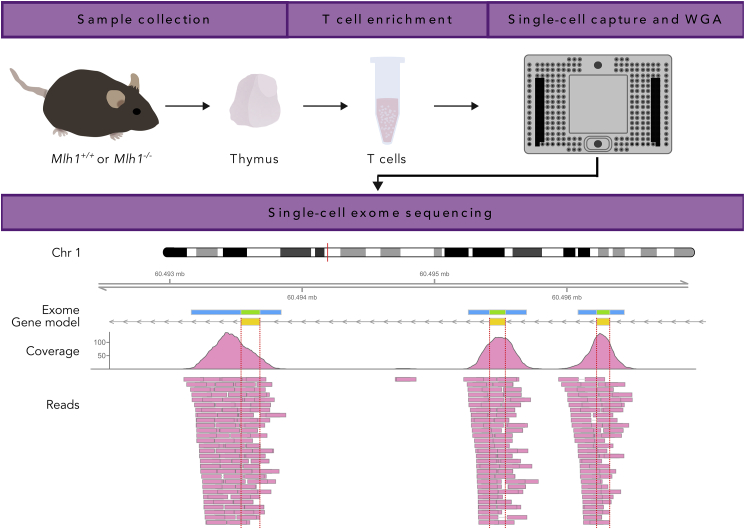
Figure 1.
Whole-Exome Sequencing of Single T Cells: Experimental Overview
Thymi of Mlh1−/− and Mlh1+/+ mice were dissected and used for enrichment of naive T cells, followed by single-cell capture, cell lysis, and whole-genome amplification in a Fluidigm C1. Amplified genomes were used for whole-exome sequencing (WES), and sequencing reads were analyzed for genetic variants. Shown is a read pileup and coverage of sample WT1-C26 in a ~5-kb-long region on chromosome 1 that contains three exons of Raph1. In addition to exons (green bar in exome panel), WES also partially covers non-coding regions adjacent to exons (blue bar in exome panel), enabling the comparison of mutation frequency between exonic and non-coding regions.