Figure 2.
Global and Local Mutation Frequencies in Single T cells
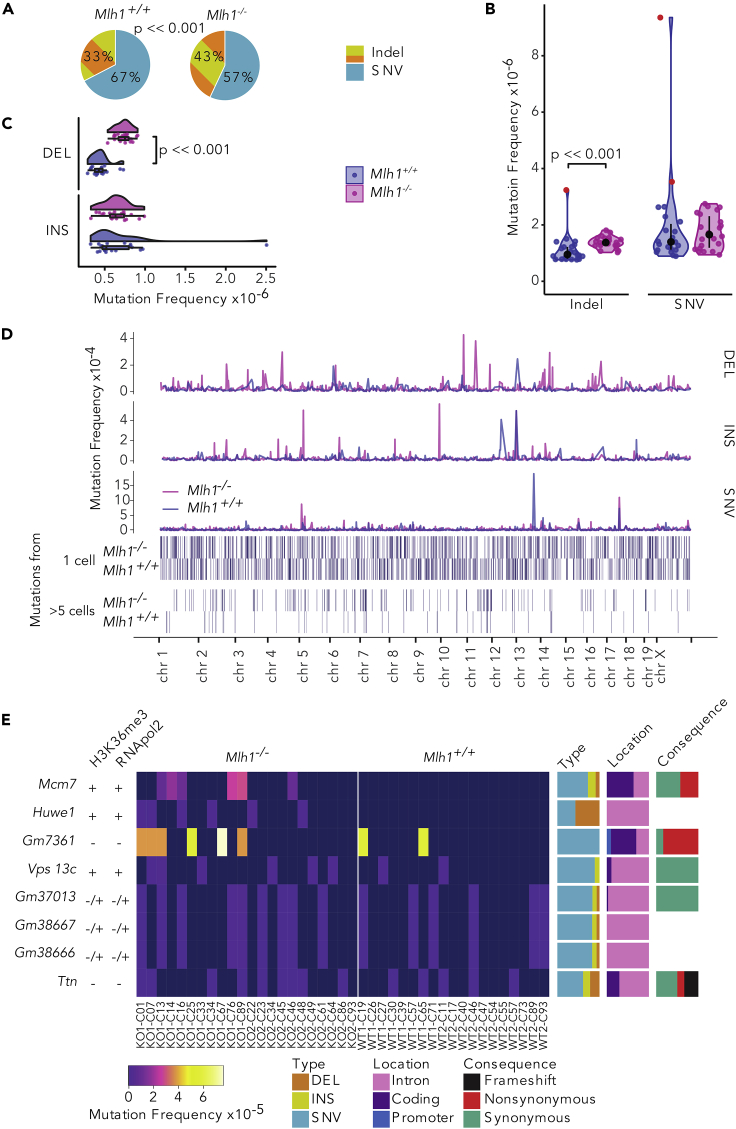
(A–C) (A) Mlh1−/− T cells have an increased amount of indels out of total mutations in the whole exome compared with Mlh1+/+ Tcells (p << 0.0001, Fisher's exact test). Global (B) indel and SNV frequencies and (C) deletion and insertion frequencies, in Mlh1+/+ and Mlh1−/− T cells.Mlh1−/− T cells have significantly higher indel, and especially deletion, frequencies than Mlh1+/+ T cells (p << 0.001, two-tailed Mann-Whitney U test). Data in (B) is shown as boxplots together with kernel probability density and individual datapoints, and data in (C) is shown as median and interquartile range together with kernel probability density and individual datapoints. Outlier cells (see Transparent Methods) section ""Outlier cells in single-cell exomes) are marked with red color in (B).
(D) Mutation frequencies in 1-Mb windows across the mouse genome. Mlh1−/− T cells have multiple high local mutation peaks originating from only a single T cell.
(E) Mcm7 and Huwe1 are mutational hotspots in Mlh1−/− T cells. Columns are sorted by genotype and cell ID (outliers excluded), rows based on the average mutation frequency. Mlh1+/+ cells have label WT and Mlh1−/− cells have label KO, biological replicates are marked with 1 and 2. Each cell has a cell identifier that originates from the Fluidigm C1 plate capture site.
Bar plots on the right show proportions of mutation types, locations, and consequences in genes. Left-hand-side columns show positivity or negativity for RNApol2 and H3K36me3 peaks (See also Figure S3).