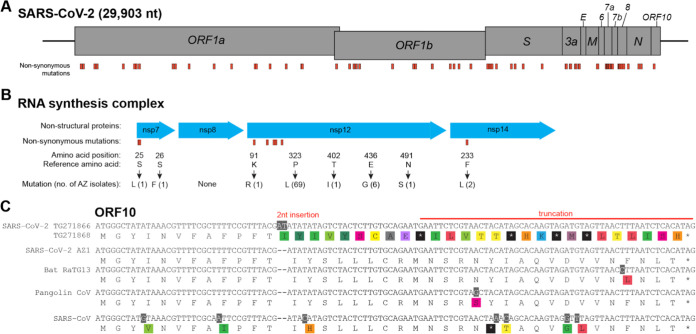
FIG 5.
Nonsynonymous mutations in Arizona isolates. (A) Diagram showing the SARS-CoV-2 genome and annotated open reading frames. The genome positions of nonsynonymous mutations in Arizona SARS-CoV-2 isolates are indicated in orange. (B) Nonsynonymous mutations of Arizona isolates in nsp’s involved in the SARS-CoV-2 RNA synthesis complex. Mutations (indicated in orange) are labeled by amino acid position within the protein, reference amino acid, amino acid change, and number of Arizona isolates with the mutation. (C) ORF10 alignment showing a 2-nucleotide insertion and subsequent early truncation in two Arizona SARS-CoV-2 isolates. GenBank and GISAID accession numbers: SARS-CoV-2 AZ-TG271866 (EPI_ISL_427271), SARS-CoV-2 AZ-TG271868 (EPI_ISL_427272), SARS-CoV-2 AZ1 (MN997409.1, EPI_ISL_406223), Bat-RaTG13 (MN996532.1), Pangolin (EPI_ISL_410721), and SARS-CoV (NC_004718.3).