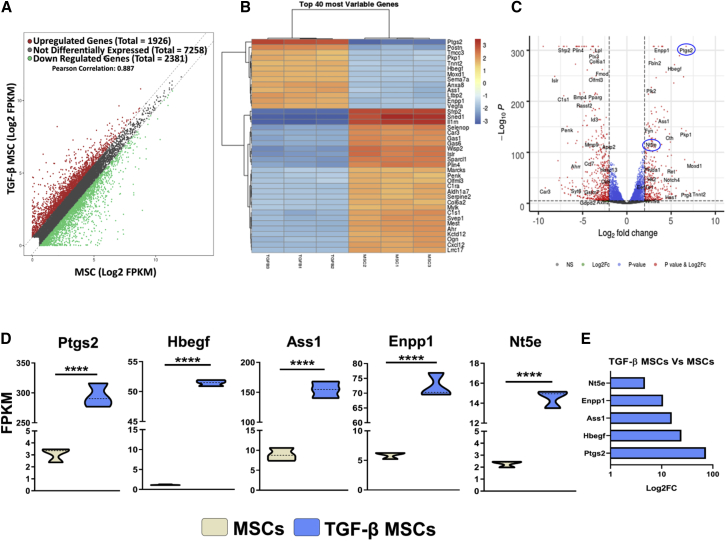
Figure 4.
Gene Expression Profile of TGF-β MSCs Compared to Untreated MSCs
MSCs were pre-treated with TGF-β for 72 h or untreated as detailed previously. RNA was isolated and RNA sequencing was performed as detailed in the Materials and Methods. (A) Scatterplot of differentially expressed genes. The scatterplot was generated by comparing genes expressed by MSCs with TGF-β MSCs with log2-scaled fragments per kilobase of transcript per million mapped reads (FPKM) values. (B) Heatmap highlighting the 40 most differentially expressed genes between MSCs (n = 3) and TGF-β MSCs (n = 3). The changing intensity of color refers to how upregulated (red) or downregulated (blue) the various genes are. (C) Volcano plot of upregulated or downregulated genes between MSCs and TGF-β MSCs, with genes involved in PGE2 synthesis and CD73 expression highlighted. (D) Violin plots showing differential FPKM for Ptgs2, Hbegf, Ass1, Enpp1, and Nt5e. Changes in FPKM values were observed for Ptgs2, Hbegf, Ass1, Enpp1, and Nt5e between MSCs and TGF-β MSCs. Dashed lines indicate median values. ∗∗∗∗p< 0.0001, by unpaired two-tailed t test. (E) Log2 fold changes for Ptgs2, Hbegf, Ass1, Enpp1, and Nt5e between MSCs and TGF-β MSCs.