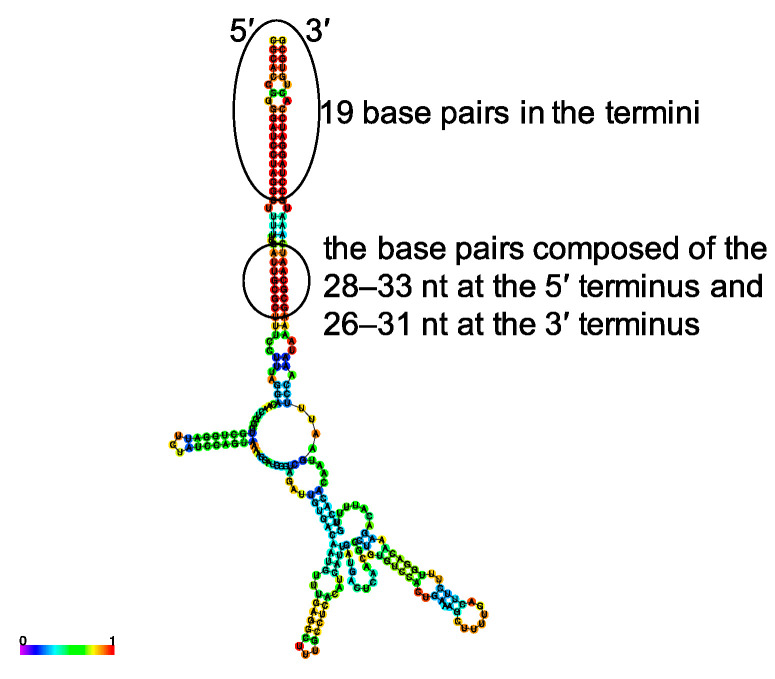
Figure 2.
Prediction of RNA secondary structures of LCMV strain WE (LCMV-WE) S segment UTR: The predicted RNA secondary structure of LCMV-WE S segment UTR derived from pRF-WE-SRG. The location of the 19 base pairs in the termini and the base pairs comprising the 28th–33rd nt in the 5′ terminus and the 26th–31st nt in the 3′ terminus are shown. RNA sequences of LCMV-WE S segment genome 5′-terminal and 3′-terminal UTRs and 50 nt of open reading frame (ORF) regional RNA sequences that were directly downstream of the UTRs were linked, sent to the CENTROIDFOLD server, and analyzed using the CONTRAfold model (weight of base pairs: 22). Each predicted base pair is colored with heat-color gradation from blue to red, corresponding to the base-pairing probability from 0 to 1. The labels “5” and “3” indicate RNA 5′ and 3′ termini, respectively.