Figure 1. Knockdown of Drosha leads to novel non-canonical microRNAs in gastric cancer cells.
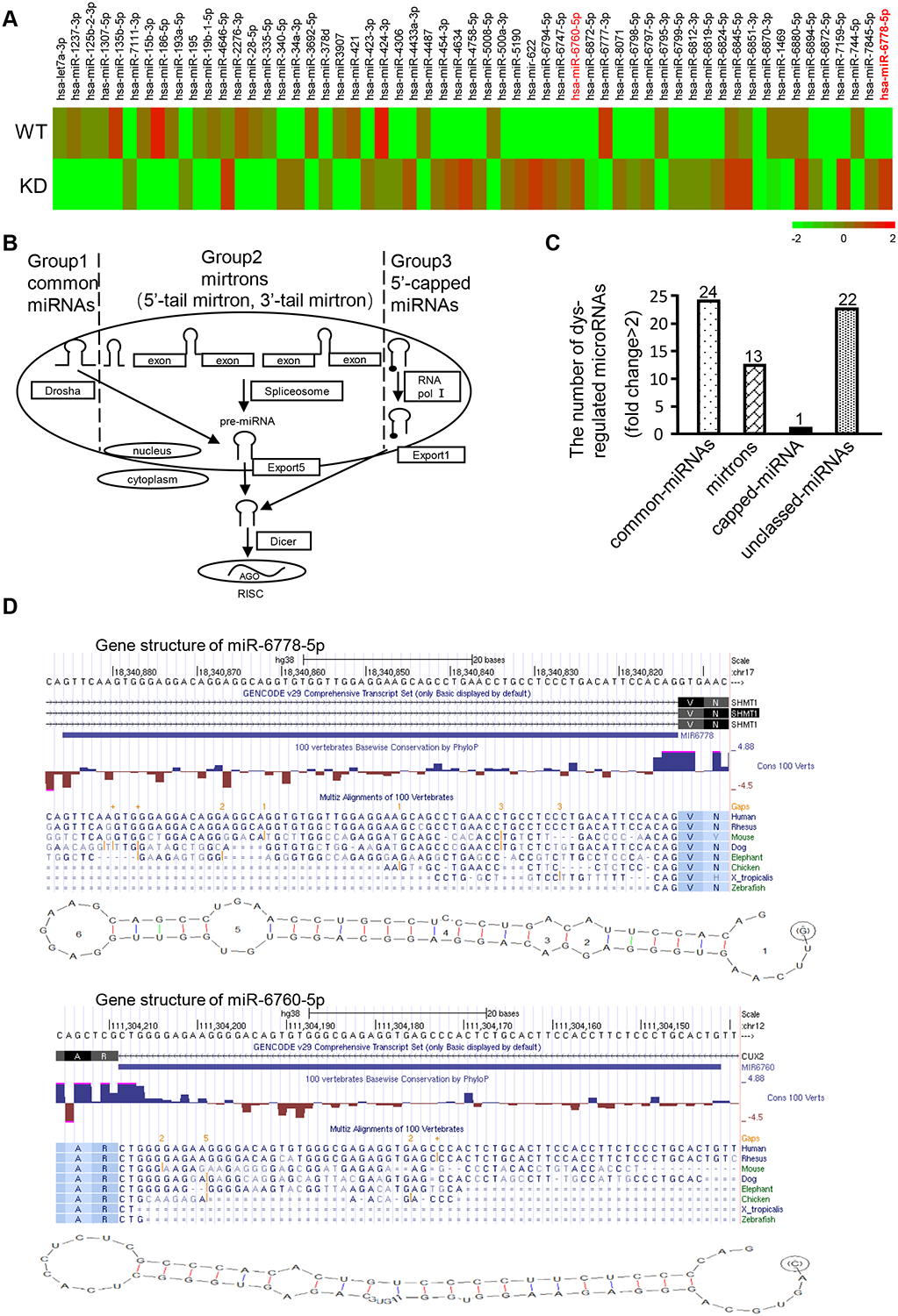
(A) A heatmap of aberrant miRNAs identified by a miRNA array of Drosha-knockdown cells. (B) Schematic diagram of canonical and splicing-mediated microRNA (miRNA) biogenesis. (C) Classification of the aberrant miRNAs in Drosha-knocked down gastric cancer cells using bioinformatics analysis. (D) Structure of a 5’ tailed mirtron. The predicted secondary structures of the investigated pre-miR-6778–5p and a known 5’ tailed mirtron (pre-miR-6760–5p) are shown. The sequences of 5’ tailed mirtrons were compared with those of their host genes, SHMT1 and CUX2, using RNA structure 5.7 software and the UCSC database (http://genome.ucsc.edu/). Human SHMT1 (miR-6778–5p) and and human CUX2 (miR-6760–5p) have the typical pyrimidine-rich region in the terminal AG splice acceptor area; the 3’ end of their pre-miRNA matches the splice acceptor in their introns.
