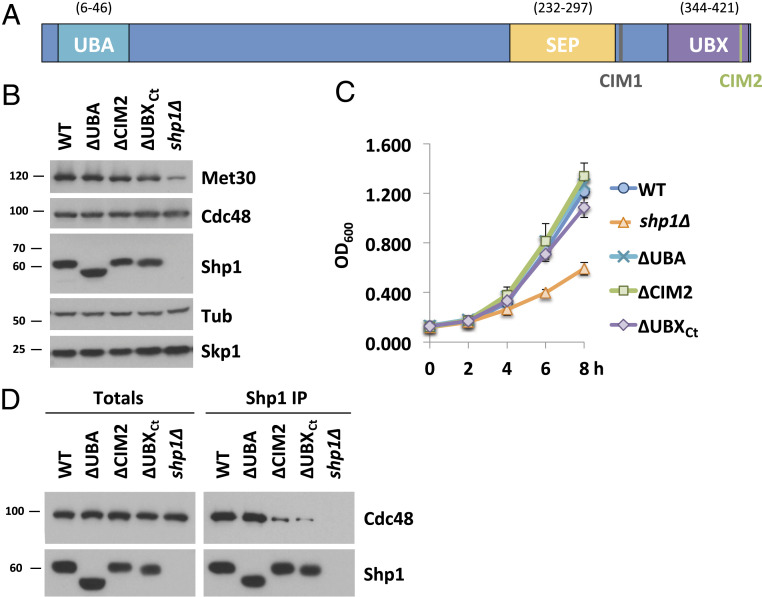
Fig. 2.
Characterization of functional domains in Shp1. (A) Schematic of Shp1 and its known functional domains and motifs. UBA, ubiquitin-associated domain (6 to 46); SEP, Shp1, eyeless and p47 domain (238 to 313); UBX, ubiquitin regulatory X domain (346 to 420); CIM 1, Cdc48-interacting motif 1 (LGGFSGQGQRL; 304 to 314 distal of SEP domain, also known as BS1); CIM2, Cdc48-interacting motif 1 (FPI; 396 to 398 in the UBX domain). (B) Expression levels of Shp1 mutants. Steady-state levels of indicated proteins were compared from strains expressing endogenous 12xMycMet30, Cdc48RGS6H, Skp1, and the various Shp1 mutants as indicated (all 3×HA-tagged). ∆UBA, deletion of residues 1 to 50; ∆CIM2, 396 to 398 FPI residues were replaced with GAG; ∆UBXCt, deletion of aa 401 to 423 or shp1 gene knocked out (K.O.). Proteins were analyzed by Western blotting with tubulin as a loading control. (C) Shp1 mutants do not show a significant growth defect at 30 °C. Wild-type cells, shp1 mutant variants as indicated, and shp1∆ (K.O.) strains were grown at 30 °C in YEPD medium, and samples were taken at indicated time points to measure optic density at 600 nm. (D) Cdc48 binding is significantly decreased in ∆CIM2 and ∆UBXCt Shp1 mutants. SHP13xHA variants were immunoprecipitated, and coprecipitation of Cdc48RGS6H was analyzed by Western blotting. The Shp1 K.O. strain was used as a background control. Results shown in B and D are representative blots from three independent experiments.